User:Vramasub/sandbox


A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions in order to stabilize the fold. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, the secondary structures underlying the zinc finger are variable and differ from protein to protein. Proteins that contain zinc fingers (zinc finger proteins) can be classified into several different structural families depending on the ligands that coordinate the zinc ion. While such proteins exist in great variety, the vast majority typically function as interaction modules that bind DNA, RNA, proteins, or other small, useful molecules. The name "zinc finger" was originally coined to describe the finger-like appearance of a diagram showing the hypothesized structure of the repeated unit in Xenopus laevis transcription factor IIIA.[1]
History[edit]
Zinc fingers were first identified in a study of transcription in the African clawed frog, Xenopus laevis. A study of the transcription of a particular RNA sequence revealed that the binding strength of a small transcription factor (transcription factor IIIA) was due to the presence of zinc-coordinating finger-like structures [2]. The name "zinc finger" was proposed in the subsequent paper, in which a detailed study of this structure alone was conducted [3]. More recent work in the characterization of proteins in various organisms has revealed the ubiquity of zinc ions in polypeptide stabilization [citation needed].
Classes[edit]
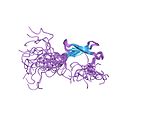
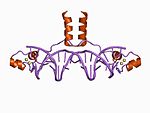
Initially, the term zinc finger was used solely to describe DNA binding motif found in Xenopus laevis, however it is now used to refer to any number of structures related by their coordination of a zinc ion. In general, zinc fingers coordinate zinc ions with a combination of cysteine and histidine residues. Originally, the number and order of these residues was used to classify different types of zinc fingers ( e.g., Cys2His2, Cys4, and Cys6). More recently, a more systematic method has been used to classify zinc finger proteins instead. This method classifies zinc finger proteins into "fold groups" based on the overall shape of the protein backbone in the folded domain. The most common "fold groups" of zinc fingers are the Cys2His2-like (the "classic zinc finger"), treble clef, and zinc ribbon.[4]
The following table[4] shows the different structures and their key features:
Cys2His2[edit]
The Cys2His2-like fold group is by far the best-characterized class of zinc fingers and are extremely common in mammalian transcription factors. These domains adopt a simple ββα fold and have the amino acid Sequence motif:[5]
X2-Cys-X2,4-Cys-X12-His-X3,4,5-His
This class of zinc fingers can have a variety of functions such as binding RNA and mediating protein-protein interactions, but is best known for its role in sequence-specific DNA-binding proteins such as Zif268. In such proteins, individual zinc finger domains typically occur as tandem repeats with two, three, or more fingers comprising the DNA-binding domain of the protein. These tandem arrays can bind in the major groove of DNA and are typically spaced at 3-bp intervals. The α-helix of each domain (often called the "recognition helix") can make sequence-specific contacts to DNA bases; residues from a single recognition helix can contact 4 or more bases to yield an overlapping pattern of contacts with adjacent zinc fingers.
Gag-knuckle[edit]
| Zinc knuckle | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | zf-CCHC | ||||||||||
| Pfam | PF00098 | ||||||||||
| InterPro | IPR001878 | ||||||||||
| SMART | SM00343 | ||||||||||
| PROSITE | PS50158 | ||||||||||
| |||||||||||
This fold group is defined by two short β-strands connected by a turn (zinc knuckle) followed by a short helix or loop and resembles the classical Cys2His2 motif with a large portion of the helix and β-hairpin truncated.
The retroviral nucleocapsid (NC) protein from HIV and other related retroviruses are examples of proteins possessing these motifs. The gag-knuckle zinc finger in the HIV NC protein is the target of a class of drugs known as zinc finger inhibitors.
Treble-clef[edit]
The treble-clef motif consists of a β-hairpin at the N-terminus and an α-helix at the C-terminus that each contribute two ligands for zinc binding, although a loop and a second β-hairpin of varying length and conformation can be present between the N-terminal β-hairpin and the C-terminal α-helix. These fingers are present in a diverse group of proteins that frequently do not share sequence or functional similarity with each other. The best-characterized proteins containing treble-clef zinc fingers are the nuclear hormone receptors.
Zinc Ribbon[edit]
| TFIIB zinc-binding | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | TF_Zn_Ribbon | ||||||||||
| Pfam | PF08271 | ||||||||||
| Pfam clan | Zn_Beta_Ribbon | ||||||||||
| InterPro | IPR013137 | ||||||||||
| PROSITE | PS51134 | ||||||||||
| |||||||||||
The zinc ribbon fold is characterised by two beta-hairpins forming two structurally similar zinc-binding sub-sites.
Zn2/Cys6[edit]
| Fungal Zn(2)-Cys(6) binuclear cluster domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | Zn_clus | ||||||||||
| Pfam | PF00172 | ||||||||||
| InterPro | IPR001138 | ||||||||||
| SMART | GAL4 | ||||||||||
| PROSITE | PS00463 | ||||||||||
| CDD | cd00067 | ||||||||||
| |||||||||||
The canonical members of this class contain a binuclear zinc cluster in which two zinc ions are bound by six cysteine residues. These zinc fingers can be found in several transcription factors including the yeast Gal4 protein.
Engineered Zinc Finger Proteins[edit]
Zinc fingers most commonly function as interaction modules; in particular, a large number have been identified as transcription factors (e.g. EGR1). As such, engineered zinc finger proteins have a variety of uses by combining the binding capacity of zinc fingers with other functional polypeptides. Such chimeric proteins, which are called zinc finger chimeras, provide powerful tools for genetic manipulation and control. The most important examples of these zinc finger chimeras are zinc finger nucleases and zinc finger transcription factors
See also[edit]
- Zinc finger nuclease
- Zinc finger inhibitor
- Steroid hormone receptor
- DNA-binding protein
- Sequence motif
- Structural motif
- RING finger domain
- B-box zinc finger
References[edit]
- ^ Klug, A and Rhodes, D (1987). "Zinc fingers: a novel protein fold for nucleic acid recognition". Cold Spring Harbor Symposia on Quantitative Biology. 52: 473–82. doi:10.1101/SQB.1987.052.01.054. PMID 3135979.
{{cite journal}}: Unknown parameter|name=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Miller J, McLachlan AD, Klug A (June 1985). "Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes". EMBO J. 4 (6): 1609–14. doi:10.1002/j.1460-2075.1985.tb03825.x. PMC 554390. PMID 4040853.
{{cite journal}}: CS1 maint: date and year (link) CS1 maint: multiple names: authors list (link) - ^ Cite error: The named reference
novel foldwas invoked but never defined (see the help page). - ^ a b Krishna SS, Majumdar I, Grishin NV (January 2003). "Structural classification of zinc fingers: survey and summary". Nucleic Acids Res. 31 (2): 532–50. doi:10.1093/nar/gkg161. PMC 140525. PMID 12527760.
{{cite journal}}: CS1 maint: date and year (link) CS1 maint: multiple names: authors list (link) - ^ C.O. Pabo; E.Peisach; R.A. Grant (2001). "Design and Selection of Novel Cys2His2 Zinc Finger Proteins". Annu. Rev. Biochem. 70: 313–40. doi:10.1146/annurev.biochem.70.1.313. PMID 11395410.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links[edit]
- McDowall J. "Protein of the Month: Zinc Fingers". European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI). Retrieved 2008-01-13.
- Goodsell DS. "Molecule of the Month: Zinc Fingers". Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). Retrieved 2008-01-13.
- The double helix between the zinc finger
- Zinc Finger Tools design and information site
- Human KZNF Gene Catalog
- Zinc finger C2H2-type domain in PROSITE
- Entry for zinc finger class C2H2 in the SMART database
- The Zinc Finger Consortium
- ZiFiT- Zinc Finger Design Tool
- Zinc Finger Consortium Materials from Addgene