Log page index: User:ProteinBoxBot/PBB_Log_Index
Protein Status Quick Log - Date: 19:00, 8 November 2007 (UTC)[edit]
Proteins without matches (21)[edit]
Proteins with a High Potential Match (4)[edit]
Created (9)[edit]
Manual Inspection (Page not found) (16)[edit]
Protein Status Grid - Date: 19:00, 8 November 2007 (UTC)[edit]
Vebose Log - Date: 19:00, 8 November 2007 (UTC)[edit]
- INFO: Beginning work on CASP10... {November 8, 2007 10:25:28 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:26:47 AM PST}
- CREATED: Created new protein page: CASP10 {November 8, 2007 10:26:56 AM PST}
- INFO: Beginning work on CYP11A1... {November 8, 2007 10:26:56 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:27:36 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Cytochrome P450, family 11, subfamily A, polypeptide 1
| HGNCid = 2590
| Symbol = CYP11A1
| AltSymbols =; CYP11A; P450SCC
| OMIM = 118485
| ECnumber =
| Homologene = 37347
| MGIid = 88582
| GeneAtlas_image1 = PBB_GE_CYP11A1_204309_at_tn.png
| Function = {{GNF_GO|id=GO:0004497 |text = monooxygenase activity}} {{GNF_GO|id=GO:0005506 |text = iron ion binding}} {{GNF_GO|id=GO:0008386 |text = cholesterol monooxygenase (side-chain-cleaving) activity}} {{GNF_GO|id=GO:0019825 |text = oxygen binding}} {{GNF_GO|id=GO:0020037 |text = heme binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}}
| Component = {{GNF_GO|id=GO:0005739 |text = mitochondrion}} {{GNF_GO|id=GO:0016020 |text = membrane}}
| Process = {{GNF_GO|id=GO:0006118 |text = electron transport}} {{GNF_GO|id=GO:0006629 |text = lipid metabolic process}} {{GNF_GO|id=GO:0006700 |text = C21-steroid hormone biosynthetic process}} {{GNF_GO|id=GO:0006702 |text = androgen biosynthetic process}} {{GNF_GO|id=GO:0008202 |text = steroid metabolic process}} {{GNF_GO|id=GO:0008203 |text = cholesterol metabolic process}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 1583
| Hs_Ensembl = ENSG00000140459
| Hs_RefseqProtein = NP_000772
| Hs_RefseqmRNA = NM_000781
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 15
| Hs_GenLoc_start = 72417193
| Hs_GenLoc_end = 72447134
| Hs_Uniprot = P05108
| Mm_EntrezGene = 13070
| Mm_Ensembl = ENSMUSG00000032323
| Mm_RefseqmRNA = XM_976231
| Mm_RefseqProtein = XP_981325
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 9
| Mm_GenLoc_start = 57813150
| Mm_GenLoc_end = 57825158
| Mm_Uniprot = Q6NV84
}}
}}
'''Cytochrome P450, family 11, subfamily A, polypeptide 1''', also known as '''CYP11A1''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This protein localizes to the mitochondrial inner membrane and catalyzes the conversion of cholesterol to pregnenolone, the first and rate-limiting step in the synthesis of the steroid hormones.<ref>{{cite web | title = Entrez Gene: CYP11A1 cytochrome P450, family 11, subfamily A, polypeptide 1| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1583| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Helmberg A |title=Twin genes and endocrine disease: CYP21 and CYP11B genes. |journal=Acta Endocrinol. |volume=129 |issue= 2 |pages= 97-108 |year= 1993 |pmid= 8372604 |doi= }}
*{{cite journal | author=Papadopoulos V, Amri H, Boujrad N, ''et al.'' |title=Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. |journal=Steroids |volume=62 |issue= 1 |pages= 21-8 |year= 1997 |pmid= 9029710 |doi= }}
*{{cite journal | author=Stocco DM |title=Intramitochondrial cholesterol transfer. |journal=Biochim. Biophys. Acta |volume=1486 |issue= 1 |pages= 184-97 |year= 2000 |pmid= 10856721 |doi= }}
*{{cite journal | author=Kristensen VN, Kure EH, Erikstein B, ''et al.'' |title=Genetic susceptibility and environmental estrogen-like compounds. |journal=Mutat. Res. |volume=482 |issue= 1-2 |pages= 77-82 |year= 2001 |pmid= 11535251 |doi= }}
*{{cite journal | author=Strauss JF |title=Some new thoughts on the pathophysiology and genetics of polycystic ovary syndrome. |journal=Ann. N. Y. Acad. Sci. |volume=997 |issue= |pages= 42-8 |year= 2004 |pmid= 14644808 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on DEK... {November 8, 2007 10:48:54 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein DEK image.jpg {November 8, 2007 10:49:28 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:49:38 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_DEK_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1q1v.
| PDB = {{PDB2|1q1v}}
| Name = DEK oncogene (DNA binding)
| HGNCid = 2768
| Symbol = DEK
| AltSymbols =; D6S231E
| OMIM = 125264
| ECnumber =
| Homologene = 2593
| MGIid = 1926209
| GeneAtlas_image1 = PBB_GE_DEK_200934_at_tn.png
| Function = {{GNF_GO|id=GO:0003677 |text = DNA binding}} {{GNF_GO|id=GO:0003704 |text = specific RNA polymerase II transcription factor activity}} {{GNF_GO|id=GO:0003723 |text = RNA binding}} {{GNF_GO|id=GO:0005525 |text = GTP binding}} {{GNF_GO|id=GO:0042393 |text = histone binding}}
| Component = {{GNF_GO|id=GO:0005634 |text = nucleus}} {{GNF_GO|id=GO:0005786 |text = signal recognition particle, endoplasmic reticulum targeting}}
| Process = {{GNF_GO|id=GO:0006357 |text = regulation of transcription from RNA polymerase II promoter}} {{GNF_GO|id=GO:0006614 |text = SRP-dependent cotranslational protein targeting to membrane}} {{GNF_GO|id=GO:0007165 |text = signal transduction}} {{GNF_GO|id=GO:0019079 |text = viral genome replication}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 7913
| Hs_Ensembl = ENSG00000124795
| Hs_RefseqProtein = NP_003463
| Hs_RefseqmRNA = NM_003472
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 6
| Hs_GenLoc_start = 18332392
| Hs_GenLoc_end = 18372750
| Hs_Uniprot = P35659
| Mm_EntrezGene = 110052
| Mm_Ensembl = ENSMUSG00000021377
| Mm_RefseqmRNA = NM_025900
| Mm_RefseqProtein = NP_080176
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 13
| Mm_GenLoc_start = 47095756
| Mm_GenLoc_end = 47117138
| Mm_Uniprot = Q3U4W4
}}
}}
'''DEK oncogene (DNA binding)''', also known as '''DEK''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a protein with one SAP domain. This protein binds to cruciform and superhelical DNA and induces positive supercoils into closed circular DNA and is also involved in splice site selection during mRNA processing. Chromosomal aberrations involving this region, increased expression of this gene and the presence of antibodies against this protein are all associated with various diseases.<ref>{{cite web | title = Entrez Gene: DEK DEK oncogene (DNA binding)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7913| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=von Lindern M, Fornerod M, Soekarman N, ''et al.'' |title=Translocation t(6;9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. |journal=Baillieres Clin. Haematol. |volume=5 |issue= 4 |pages= 857-79 |year= 1993 |pmid= 1308167 |doi= }}
*{{cite journal | author=Waldmann T, Scholten I, Kappes F, ''et al.'' |title=The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. |journal=Gene |volume=343 |issue= 1 |pages= 1-9 |year= 2005 |pmid= 15563827 |doi= 10.1016/j.gene.2004.08.029 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on FASN... {November 8, 2007 10:29:16 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein FASN image.jpg {November 8, 2007 10:30:09 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:30:28 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_FASN_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1xkt.
| PDB = {{PDB2|1xkt}}, {{PDB2|2cg5}}, {{PDB2|2jfd}}, {{PDB2|2jfk}}
| Name = Fatty acid synthase
| HGNCid = 3594
| Symbol = FASN
| AltSymbols =; FAS; MGC14367; MGC15706; OA-519
| OMIM = 600212
| ECnumber =
| Homologene = 55800
| MGIid = 95485
| GeneAtlas_image1 = PBB_GE_FASN_212218_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_FASN_217006_x_at_tn.png
| Function = {{GNF_GO|id=GO:0003824 |text = catalytic activity}} {{GNF_GO|id=GO:0004313 |text = [acyl-carrier-protein] S-acetyltransferase activity}} {{GNF_GO|id=GO:0004314 |text = [acyl-carrier-protein] S-malonyltransferase activity}} {{GNF_GO|id=GO:0004315 |text = 3-oxoacyl-[acyl-carrier-protein] synthase activity}} {{GNF_GO|id=GO:0004316 |text = 3-oxoacyl-[acyl-carrier-protein] reductase activity}} {{GNF_GO|id=GO:0004317 |text = 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase activity}} {{GNF_GO|id=GO:0004319 |text = enoyl-[acyl-carrier-protein] reductase (NADPH, B-specific) activity}} {{GNF_GO|id=GO:0004320 |text = oleoyl-[acyl-carrier-protein] hydrolase activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0010281 |text = acyl-ACP thioesterase activity}} {{GNF_GO|id=GO:0016491 |text = oxidoreductase activity}} {{GNF_GO|id=GO:0016740 |text = transferase activity}} {{GNF_GO|id=GO:0016788 |text = hydrolase activity, acting on ester bonds}} {{GNF_GO|id=GO:0016829 |text = lyase activity}} {{GNF_GO|id=GO:0019901 |text = protein kinase binding}} {{GNF_GO|id=GO:0031177 |text = phosphopantetheine binding}} {{GNF_GO|id=GO:0048037 |text = cofactor binding}}
| Component = {{GNF_GO|id=GO:0042587 |text = glycogen granule}}
| Process = {{GNF_GO|id=GO:0006633 |text = fatty acid biosynthetic process}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 2194
| Hs_Ensembl = ENSG00000169710
| Hs_RefseqProtein = NP_004095
| Hs_RefseqmRNA = NM_004104
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 17
| Hs_GenLoc_start = 77629504
| Hs_GenLoc_end = 77649395
| Hs_Uniprot = P49327
| Mm_EntrezGene = 14104
| Mm_Ensembl = ENSMUSG00000025153
| Mm_RefseqmRNA = NM_007988
| Mm_RefseqProtein = NP_032014
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 11
| Mm_GenLoc_start = 120623167
| Mm_GenLoc_end = 120640332
| Mm_Uniprot = O89060
}}
}}
'''Fatty acid synthase''', also known as '''FASN''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = The enzyme encoded by this gene is a multifunctional protein. Its main function is to catalyze the synthesis of palmitate from acetyl-CoA and malonyl-CoA, in the presence of NADPH, into long-chain saturated fatty acids. In some cancer cell lines, this protein has been found to be fused with estrogen receptor-alpha (ER-alpha), in which the N-terminus of FAS is fused in-frame with the C-terminus of ER-alpha.<ref>{{cite web | title = Entrez Gene: FASN fatty acid synthase| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=2194| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Wakil SJ |title=Fatty acid synthase, a proficient multifunctional enzyme. |journal=Biochemistry |volume=28 |issue= 11 |pages= 4523-30 |year= 1989 |pmid= 2669958 |doi= }}
*{{cite journal | author=Baron A, Migita T, Tang D, Loda M |title=Fatty acid synthase: a metabolic oncogene in prostate cancer? |journal=J. Cell. Biochem. |volume=91 |issue= 1 |pages= 47-53 |year= 2004 |pmid= 14689581 |doi= 10.1002/jcb.10708 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on FBLN1... {November 8, 2007 10:27:36 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:29:09 AM PST}
- CREATED: Created new protein page: FBLN1 {November 8, 2007 10:29:16 AM PST}
- INFO: Beginning work on FCGR1A... {November 8, 2007 10:30:28 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:31:30 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Fc fragment of IgG, high affinity Ia, receptor (CD64)
| HGNCid = 3613
| Symbol = FCGR1A
| AltSymbols =; CD64; CD64A; FCRI; IGFR1
| OMIM = 146760
| ECnumber =
| Homologene = 475
| MGIid = 95498
| GeneAtlas_image1 = PBB_GE_FCGR1A_216950_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_FCGR1A_214511_x_at_tn.png
| Function = {{GNF_GO|id=GO:0004872 |text = receptor activity}} {{GNF_GO|id=GO:0005057 |text = receptor signaling protein activity}} {{GNF_GO|id=GO:0019864 |text = IgG binding}}
| Component = {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}}
| Process = {{GNF_GO|id=GO:0006911 |text = phagocytosis, engulfment}} {{GNF_GO|id=GO:0006955 |text = immune response}} {{GNF_GO|id=GO:0007165 |text = signal transduction}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 2209
| Hs_Ensembl = ENSG00000150337
| Hs_RefseqProtein = NP_000557
| Hs_RefseqmRNA = NM_000566
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 1
| Hs_GenLoc_start = 148020870
| Hs_GenLoc_end = 148030698
| Hs_Uniprot = P12314
| Mm_EntrezGene = 14129
| Mm_Ensembl = ENSMUSG00000015947
| Mm_RefseqmRNA = NM_010186
| Mm_RefseqProtein = NP_034316
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 3
| Mm_GenLoc_start = 96368314
| Mm_GenLoc_end = 96379374
| Mm_Uniprot = P26151
}}
}}
'''Fc fragment of IgG, high affinity Ia, receptor (CD64)''', also known as '''FCGR1A''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Monteiro RC, Van De Winkel JG |title=IgA Fc receptors. |journal=Annu. Rev. Immunol. |volume=21 |issue= |pages= 177-204 |year= 2003 |pmid= 12524384 |doi= 10.1146/annurev.immunol.21.120601.141011 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on FECH... {November 8, 2007 10:31:30 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein FECH image.jpg {November 8, 2007 10:32:18 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:32:31 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_FECH_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1hrk.
| PDB = {{PDB2|1hrk}}, {{PDB2|2hrc}}, {{PDB2|2hre}}
| Name = Ferrochelatase (protoporphyria)
| HGNCid = 3647
| Symbol = FECH
| AltSymbols =; EPP; FCE
| OMIM = 177000
| ECnumber =
| Homologene = 113
| MGIid = 95513
| GeneAtlas_image1 = PBB_GE_FECH_203116_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_FECH_203115_at_tn.png
| Function = {{GNF_GO|id=GO:0004325 |text = ferrochelatase activity}} {{GNF_GO|id=GO:0008198 |text = ferrous iron binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}} {{GNF_GO|id=GO:0051537 |text = 2 iron, 2 sulfur cluster binding}}
| Component = {{GNF_GO|id=GO:0005739 |text = mitochondrion}}
| Process = {{GNF_GO|id=GO:0006091 |text = generation of precursor metabolites and energy}} {{GNF_GO|id=GO:0006783 |text = heme biosynthetic process}} {{GNF_GO|id=GO:0009416 |text = response to light stimulus}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 2235
| Hs_Ensembl = ENSG00000066926
| Hs_RefseqProtein = NP_000131
| Hs_RefseqmRNA = NM_000140
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 18
| Hs_GenLoc_start = 53366535
| Hs_GenLoc_end = 53404988
| Hs_Uniprot = P22830
| Mm_EntrezGene = 14151
| Mm_Ensembl = ENSMUSG00000024588
| Mm_RefseqmRNA = XM_001000060
| Mm_RefseqProtein = XP_001000060
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 18
| Mm_GenLoc_start = 64581919
| Mm_GenLoc_end = 64614435
| Mm_Uniprot = Q3UC49
}}
}}
'''Ferrochelatase (protoporphyria)''', also known as '''FECH''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Ferrochelatase is localized to the mitochondrion where it catalyzes the insertion of the ferrous form of iron into protoporphyrin IX in the heme synthesis pathway. Defects in ferrochelatase are associated with protoporphyria. Two transcript variants encoding different isoforms have been found for this gene.<ref>{{cite web | title = Entrez Gene: FECH ferrochelatase (protoporphyria)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=2235| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Cox TM |title=Erythropoietic protoporphyria. |journal=J. Inherit. Metab. Dis. |volume=20 |issue= 2 |pages= 258-69 |year= 1997 |pmid= 9211198 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on GGA1... {November 8, 2007 10:50:25 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein GGA1 image.jpg {November 8, 2007 10:51:33 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:51:41 AM PST}
- CREATED: Created new protein page: GGA1 {November 8, 2007 10:51:48 AM PST}
- INFO: Beginning work on GNAI3... {November 8, 2007 10:32:31 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein GNAI3 image.jpg {November 8, 2007 10:34:14 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:34:27 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_GNAI3_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1agr.
| PDB = {{PDB2|1agr}}, {{PDB2|1as0}}, {{PDB2|1as2}}, {{PDB2|1as3}}, {{PDB2|1bh2}}, {{PDB2|1bof}}, {{PDB2|1cip}}, {{PDB2|1gdd}}, {{PDB2|1gfi}}, {{PDB2|1gg2}}, {{PDB2|1gia}}, {{PDB2|1gil}}, {{PDB2|1git}}, {{PDB2|1gp2}}, {{PDB2|1kjy}}, {{PDB2|1svk}}, {{PDB2|1svs}}, {{PDB2|1y3a}}, {{PDB2|2g83}}, {{PDB2|2gtp}}, {{PDB2|2hlb}}, {{PDB2|2ihb}}, {{PDB2|2ik8}}, {{PDB2|2ode}}
| Name = Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
| HGNCid = 4387
| Symbol = GNAI3
| AltSymbols =; 87U6; FLJ26559
| OMIM = 139370
| ECnumber =
| Homologene = 55975
| MGIid = 95773
| GeneAtlas_image1 = PBB_GE_GNAI3_201180_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_GNAI3_201179_s_at_tn.png
| GeneAtlas_image3 = PBB_GE_GNAI3_201181_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0003924 |text = GTPase activity}} {{GNF_GO|id=GO:0004871 |text = signal transducer activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005525 |text = GTP binding}}
| Component = {{GNF_GO|id=GO:0005737 |text = cytoplasm}} {{GNF_GO|id=GO:0005794 |text = Golgi apparatus}}
| Process = {{GNF_GO|id=GO:0006810 |text = transport}} {{GNF_GO|id=GO:0007165 |text = signal transduction}} {{GNF_GO|id=GO:0007186 |text = G-protein coupled receptor protein signaling pathway}} {{GNF_GO|id=GO:0007194 |text = negative regulation of adenylate cyclase activity}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 2773
| Hs_Ensembl = ENSG00000065135
| Hs_RefseqProtein = NP_006487
| Hs_RefseqmRNA = NM_006496
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 1
| Hs_GenLoc_start = 109892756
| Hs_GenLoc_end = 109938498
| Hs_Uniprot = P08754
| Mm_EntrezGene = 14679
| Mm_Ensembl = ENSMUSG00000000001
| Mm_RefseqmRNA = NM_010306
| Mm_RefseqProtein = NP_034436
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 3
| Mm_GenLoc_start = 108235337
| Mm_GenLoc_end = 108274202
| Mm_Uniprot = Q3TJH1
}}
}}
'''Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3''', also known as '''GNAI3''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Downes GB, Gautam N |title=The G protein subunit gene families. |journal=Genomics |volume=62 |issue= 3 |pages= 544-52 |year= 2000 |pmid= 10644457 |doi= 10.1006/geno.1999.5992 }}
*{{cite journal | author=Sidhu A, Niznik HB |title=Coupling of dopamine receptor subtypes to multiple and diverse G proteins. |journal=Int. J. Dev. Neurosci. |volume=18 |issue= 7 |pages= 669-77 |year= 2000 |pmid= 10978845 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on MAP2... {November 8, 2007 10:37:23 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:38:17 AM PST}
- CREATED: Created new protein page: MAP2 {November 8, 2007 10:38:24 AM PST}
- INFO: Beginning work on MYO7A... {November 8, 2007 10:38:24 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:38:46 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Myosin VIIA
| HGNCid = 7606
| Symbol = MYO7A
| AltSymbols =; DFNA11; DFNB2; MYU7A; NSRD2; USH1B
| OMIM = 276903
| ECnumber =
| Homologene = 219
| MGIid = 104510
| Function = {{GNF_GO|id=GO:0000146 |text = microfilament motor activity}} {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0003779 |text = actin binding}} {{GNF_GO|id=GO:0005488 |text = binding}} {{GNF_GO|id=GO:0005516 |text = calmodulin binding}} {{GNF_GO|id=GO:0005524 |text = ATP binding}} {{GNF_GO|id=GO:0046983 |text = protein dimerization activity}}
| Component = {{GNF_GO|id=GO:0001750 |text = photoreceptor outer segment}} {{GNF_GO|id=GO:0001917 |text = photoreceptor inner segment}} {{GNF_GO|id=GO:0005737 |text = cytoplasm}} {{GNF_GO|id=GO:0005765 |text = lysosomal membrane}} {{GNF_GO|id=GO:0005829 |text = cytosol}} {{GNF_GO|id=GO:0005856 |text = cytoskeleton}} {{GNF_GO|id=GO:0005929 |text = cilium}} {{GNF_GO|id=GO:0016324 |text = apical plasma membrane}} {{GNF_GO|id=GO:0016459 |text = myosin complex}} {{GNF_GO|id=GO:0042470 |text = melanosome}} {{GNF_GO|id=GO:0045202 |text = synapse}}
| Process = {{GNF_GO|id=GO:0001845 |text = phagolysosome formation}} {{GNF_GO|id=GO:0007040 |text = lysosome organization and biogenesis}} {{GNF_GO|id=GO:0007601 |text = visual perception}} {{GNF_GO|id=GO:0007605 |text = sensory perception of sound}} {{GNF_GO|id=GO:0030030 |text = cell projection organization and biogenesis}} {{GNF_GO|id=GO:0030048 |text = actin filament-based movement}} {{GNF_GO|id=GO:0035058 |text = sensory cilium biogenesis}} {{GNF_GO|id=GO:0042472 |text = inner ear morphogenesis}} {{GNF_GO|id=GO:0042491 |text = auditory receptor cell differentiation}} {{GNF_GO|id=GO:0048563 |text = post-embryonic organ morphogenesis}} {{GNF_GO|id=GO:0050896 |text = response to stimulus}} {{GNF_GO|id=GO:0051875 |text = pigment granule localization}} {{GNF_GO|id=GO:0051904 |text = pigment granule transport}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 4647
| Hs_Ensembl =
| Hs_RefseqProtein = NP_000251
| Hs_RefseqmRNA = NM_000260
| Hs_GenLoc_db =
| Hs_GenLoc_chr =
| Hs_GenLoc_start =
| Hs_GenLoc_end =
| Hs_Uniprot =
| Mm_EntrezGene = 17921
| Mm_Ensembl = ENSMUSG00000030761
| Mm_RefseqmRNA = NM_008663
| Mm_RefseqProtein = NP_032689
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 7
| Mm_GenLoc_start = 97926593
| Mm_GenLoc_end = 97984444
| Mm_Uniprot = P97479
}}
}}
'''Myosin VIIA''', also known as '''MYO7A''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Wolfrum U |title=The cellular function of the usher gene product myosin VIIa is specified by its ligands. |journal=Adv. Exp. Med. Biol. |volume=533 |issue= |pages= 133-42 |year= 2004 |pmid= 15180257 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on NR4A2... {November 8, 2007 10:38:46 AM PST}
- UPLOAD: Added new Image to wiki:
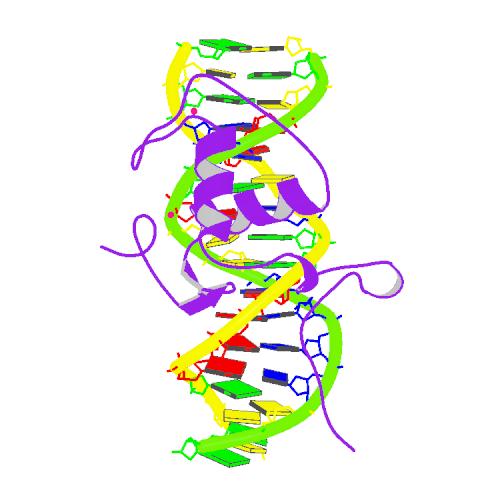 {November 8, 2007 10:39:55 AM PST}
{November 8, 2007 10:39:55 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:40:14 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_NR4A2_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1cit.
| PDB = {{PDB2|1cit}}, {{PDB2|1ovl}}
| Name = Nuclear receptor subfamily 4, group A, member 2
| HGNCid = 7981
| Symbol = NR4A2
| AltSymbols =; RNR1; HZF-3; NOT; NURR1; TINUR
| OMIM = 601828
| ECnumber =
| Homologene = 4509
| MGIid = 1352456
| GeneAtlas_image1 = PBB_GE_NR4A2_216248_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_NR4A2_204621_s_at_tn.png
| GeneAtlas_image3 = PBB_GE_NR4A2_204622_x_at_tn.png
| Function = {{GNF_GO|id=GO:0003700 |text = transcription factor activity}} {{GNF_GO|id=GO:0003707 |text = steroid hormone receptor activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0008270 |text = zinc ion binding}} {{GNF_GO|id=GO:0043565 |text = sequence-specific DNA binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}}
| Component = {{GNF_GO|id=GO:0005634 |text = nucleus}}
| Process = {{GNF_GO|id=GO:0006350 |text = transcription}} {{GNF_GO|id=GO:0006355 |text = regulation of transcription, DNA-dependent}} {{GNF_GO|id=GO:0007165 |text = signal transduction}} {{GNF_GO|id=GO:0007399 |text = nervous system development}} {{GNF_GO|id=GO:0019735 |text = antimicrobial humoral response}} {{GNF_GO|id=GO:0030182 |text = neuron differentiation}} {{GNF_GO|id=GO:0042053 |text = regulation of dopamine metabolic process}} {{GNF_GO|id=GO:0045944 |text = positive regulation of transcription from RNA polymerase II promoter}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 4929
| Hs_Ensembl = ENSG00000153234
| Hs_RefseqProtein = NP_006177
| Hs_RefseqmRNA = NM_006186
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 2
| Hs_GenLoc_start = 156889194
| Hs_GenLoc_end = 156897474
| Hs_Uniprot = P43354
| Mm_EntrezGene = 18227
| Mm_Ensembl = ENSMUSG00000026826
| Mm_RefseqmRNA = NM_013613
| Mm_RefseqProtein = NP_038641
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 2
| Mm_GenLoc_start = 56922903
| Mm_GenLoc_end = 56939527
| Mm_Uniprot = Q3TYI4
}}
}}
'''Nuclear receptor subfamily 4, group A, member 2''', also known as '''NR4A2''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a member of the steroid-thyroid hormone-retinoid receptor superfamily. The encoded protein may act as a transcription factor. Mutations in this gene have been associated with disorders related to dopaminergic dysfunction, including Parkinson disease, schizophernia, and manic depression. Misregulation of this gene may be associated with rheumatoid arthritis. Four transcript variants encoding four distinct isoforms have been identified for this gene. Additional alternate splice variants may exist, but their full length nature has not been determined.<ref>{{cite web | title = Entrez Gene: NR4A2 nuclear receptor subfamily 4, group A, member 2| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4929| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Le W, Appel SH |title=Mutant genes responsible for Parkinson's disease. |journal=Current opinion in pharmacology |volume=4 |issue= 1 |pages= 79-84 |year= 2004 |pmid= 15018843 |doi= 10.1016/j.coph.2003.09.005 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on NUP62... {November 8, 2007 10:49:38 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:50:17 AM PST}
- CREATED: Created new protein page: NUP62 {November 8, 2007 10:50:25 AM PST}
- INFO: Beginning work on PDHA1... {November 8, 2007 10:40:14 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PDHA1 image.jpg {November 8, 2007 10:41:08 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:41:22 AM PST}
- CREATED: Created new protein page: PDHA1 {November 8, 2007 10:41:29 AM PST}
- INFO: Beginning work on PHB... {November 8, 2007 10:42:05 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:42:55 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Prohibitin
| HGNCid = 8912
| Symbol = PHB
| AltSymbols =;
| OMIM = 176705
| ECnumber =
| Homologene = 1980
| MGIid = 97572
| GeneAtlas_image1 = PBB_GE_PHB_200658_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_PHB_200659_s_at_tn.png
| Function = {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0016563 |text = transcription activator activity}} {{GNF_GO|id=GO:0016564 |text = transcription repressor activity}}
| Component = {{GNF_GO|id=GO:0005634 |text = nucleus}} {{GNF_GO|id=GO:0005654 |text = nucleoplasm}} {{GNF_GO|id=GO:0005739 |text = mitochondrion}} {{GNF_GO|id=GO:0005743 |text = mitochondrial inner membrane}} {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}}
| Process = {{GNF_GO|id=GO:0000074 |text = regulation of progression through cell cycle}} {{GNF_GO|id=GO:0006260 |text = DNA replication}} {{GNF_GO|id=GO:0007165 |text = signal transduction}} {{GNF_GO|id=GO:0008285 |text = negative regulation of cell proliferation}} {{GNF_GO|id=GO:0016481 |text = negative regulation of transcription}} {{GNF_GO|id=GO:0016575 |text = histone deacetylation}} {{GNF_GO|id=GO:0042981 |text = regulation of apoptosis}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 5245
| Hs_Ensembl = ENSG00000167085
| Hs_RefseqProtein = NP_002625
| Hs_RefseqmRNA = NM_002634
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 17
| Hs_GenLoc_start = 44836419
| Hs_GenLoc_end = 44847241
| Hs_Uniprot = P35232
| Mm_EntrezGene = 18673
| Mm_Ensembl =
| Mm_RefseqmRNA = NM_008831
| Mm_RefseqProtein = NP_032857
| Mm_GenLoc_db =
| Mm_GenLoc_chr =
| Mm_GenLoc_start =
| Mm_GenLoc_end =
| Mm_Uniprot =
}}
}}
'''Prohibitin''', also known as '''PHB''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Prohibitin is an evolutionarily conserved gene that is ubiquitously expressed. It is thought to be a negative regulator of cell proliferation and may be a tumor suppressor. Mutations in PHB have been linked to sporadic breast cancer. Prohibitin is expressed as two transcripts with varying lengths of 3' untranslated region. The longer transcript is present at higher levels in proliferating tissues and cells, suggesting that this longer 3' untranslated region may function as a trans-acting regulatory RNA.<ref>{{cite web | title = Entrez Gene: PHB prohibitin| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5245| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=McClung JK, Jupe ER, Liu XT, Dell'Orco RT |title=Prohibitin: potential role in senescence, development, and tumor suppression. |journal=Exp. Gerontol. |volume=30 |issue= 2 |pages= 99-124 |year= 1996 |pmid= 8591812 |doi= }}
*{{cite journal | author=Dell'Orco RT, McClung JK, Jupe ER, Liu XT |title=Prohibitin and the senescent phenotype. |journal=Exp. Gerontol. |volume=31 |issue= 1-2 |pages= 245-52 |year= 1996 |pmid= 8706794 |doi= }}
*{{cite journal | author=Mishra S, Murphy LC, Nyomba BL, Murphy LJ |title=Prohibitin: a potential target for new therapeutics. |journal=Trends in molecular medicine |volume=11 |issue= 4 |pages= 192-7 |year= 2005 |pmid= 15823758 |doi= 10.1016/j.molmed.2005.02.004 }}
*{{cite journal | author=Rajalingam K, Rudel T |title=Ras-Raf signaling needs prohibitin. |journal=Cell Cycle |volume=4 |issue= 11 |pages= 1503-5 |year= 2007 |pmid= 16294014 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on PIM1... {November 8, 2007 10:42:55 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PIM1 image.jpg {November 8, 2007 10:43:31 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:43:41 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_PIM1_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1xqz.
| PDB = {{PDB2|1xqz}}, {{PDB2|1xr1}}, {{PDB2|1xws}}, {{PDB2|1yhs}}, {{PDB2|1yi3}}, {{PDB2|1yi4}}, {{PDB2|1ywv}}, {{PDB2|1yxs}}, {{PDB2|1yxt}}, {{PDB2|1yxu}}, {{PDB2|1yxv}}, {{PDB2|1yxx}}, {{PDB2|2bik}}, {{PDB2|2bil}}, {{PDB2|2bzh}}, {{PDB2|2bzi}}, {{PDB2|2bzj}}, {{PDB2|2bzk}}, {{PDB2|2c3i}}, {{PDB2|2j2i}}, {{PDB2|2o3p}}, {{PDB2|2o63}}, {{PDB2|2o64}}, {{PDB2|2o65}}, {{PDB2|2obj}}, {{PDB2|2oi4}}
| Name = Pim-1 oncogene
| HGNCid = 8986
| Symbol = PIM1
| AltSymbols =; PIM
| OMIM = 164960
| ECnumber =
| Homologene = 11214
| MGIid = 97584
| GeneAtlas_image1 = PBB_GE_PIM1_209193_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0004674 |text = protein serine/threonine kinase activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005524 |text = ATP binding}} {{GNF_GO|id=GO:0016740 |text = transferase activity}} {{GNF_GO|id=GO:0030145 |text = manganese ion binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}}
| Component = {{GNF_GO|id=GO:0005634 |text = nucleus}} {{GNF_GO|id=GO:0005737 |text = cytoplasm}}
| Process = {{GNF_GO|id=GO:0006468 |text = protein amino acid phosphorylation}} {{GNF_GO|id=GO:0007275 |text = multicellular organismal development}} {{GNF_GO|id=GO:0008283 |text = cell proliferation}} {{GNF_GO|id=GO:0043066 |text = negative regulation of apoptosis}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 5292
| Hs_Ensembl = ENSG00000137193
| Hs_RefseqProtein = NP_002639
| Hs_RefseqmRNA = NM_002648
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 6
| Hs_GenLoc_start = 37245957
| Hs_GenLoc_end = 37251180
| Hs_Uniprot = P11309
| Mm_EntrezGene = 18712
| Mm_Ensembl = ENSMUSG00000024014
| Mm_RefseqmRNA = XM_991765
| Mm_RefseqProtein = XP_996859
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 17
| Mm_GenLoc_start = 29217824
| Mm_GenLoc_end = 29222496
| Mm_Uniprot = Q3TYQ0
}}
}}
'''Pim-1 oncogene''', also known as '''PIM1''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = The protooncogene PIM1 encodes a protein kinase upregulated in prostate cancer.[supplied by OMIM]<ref>{{cite web | title = Entrez Gene: PIM1 pim-1 oncogene| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5292| accessdate = }}</ref>
}}
==References==
{{reflist}}
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on PRKAA2... {November 8, 2007 10:43:41 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PRKAA2 image.jpg {November 8, 2007 10:44:26 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:44:41 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_PRKAA2_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 2h6d.
| PDB = {{PDB2|2h6d}}
| Name = Protein kinase, AMP-activated, alpha 2 catalytic subunit
| HGNCid = 9377
| Symbol = PRKAA2
| AltSymbols =; AMPK; AMPK2; PRKAA
| OMIM = 600497
| ECnumber =
| Homologene = 4551
| MGIid = 1336173
| GeneAtlas_image1 = PBB_GE_PRKAA2_207709_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0000287 |text = magnesium ion binding}} {{GNF_GO|id=GO:0004674 |text = protein serine/threonine kinase activity}} {{GNF_GO|id=GO:0005524 |text = ATP binding}} {{GNF_GO|id=GO:0016740 |text = transferase activity}}
| Component = {{GNF_GO|id=GO:0005634 |text = nucleus}}
| Process = {{GNF_GO|id=GO:0006468 |text = protein amino acid phosphorylation}} {{GNF_GO|id=GO:0006633 |text = fatty acid biosynthetic process}} {{GNF_GO|id=GO:0006695 |text = cholesterol biosynthetic process}} {{GNF_GO|id=GO:0007165 |text = signal transduction}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 5563
| Hs_Ensembl = ENSG00000162409
| Hs_RefseqProtein = NP_006243
| Hs_RefseqmRNA = NM_006252
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 1
| Hs_GenLoc_start = 56883583
| Hs_GenLoc_end = 56953596
| Hs_Uniprot = P54646
| Mm_EntrezGene = 108079
| Mm_Ensembl = ENSMUSG00000028518
| Mm_RefseqmRNA = NM_178143
| Mm_RefseqProtein = NP_835279
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 4
| Mm_GenLoc_start = 104532922
| Mm_GenLoc_end = 104607801
| Mm_Uniprot = Q8BRK8
}}
}}
'''Protein kinase, AMP-activated, alpha 2 catalytic subunit''', also known as '''PRKAA2''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = The protein encoded by this gene is a catalytic subunit of the AMP-activated protein kinase (AMPK). AMPK is a heterotrimer consisting of an alpha catalytic subunit, and non-catalytic beta and gamma subunits. AMPK is an important energy-sensing enzyme that monitors cellular energy status. In response to cellular metabolic stresses, AMPK is activated, and thus phosphorylates and inactivates acetyl-CoA carboxylase (ACC) and beta-hydroxy beta-methylglutaryl-CoA reductase (HMGCR), key enzymes involved in regulating de novo biosynthesis of fatty acid and cholesterol. Studies of the mouse counterpart suggest that this catalytic subunit may control whole-body insulin sensitivity and is necessary for maintaining myocardial energy homeostasis during ischemia.<ref>{{cite web | title = Entrez Gene: PRKAA2 protein kinase, AMP-activated, alpha 2 catalytic subunit| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5563| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Hardie DG, MacKintosh RW |title=AMP-activated protein kinase--an archetypal protein kinase cascade? |journal=Bioessays |volume=14 |issue= 10 |pages= 699-704 |year= 1995 |pmid= 1365882 |doi= 10.1002/bies.950141011 }}
*{{cite journal | author=Hardie DG |title=Regulation of fatty acid and cholesterol metabolism by the AMP-activated protein kinase. |journal=Biochim. Biophys. Acta |volume=1123 |issue= 3 |pages= 231-8 |year= 1992 |pmid= 1536860 |doi= }}
*{{cite journal | author=Carling D |title=The AMP-activated protein kinase cascade--a unifying system for energy control. |journal=Trends Biochem. Sci. |volume=29 |issue= 1 |pages= 18-24 |year= 2004 |pmid= 14729328 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on PRMT1... {November 8, 2007 10:34:27 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PRMT1 image.jpg {November 8, 2007 10:35:01 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:35:17 AM PST}
- CREATED: Created new protein page: PRMT1 {November 8, 2007 10:35:24 AM PST}
- INFO: Beginning work on SH2D1A... {November 8, 2007 10:35:24 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein SH2D1A image.jpg {November 8, 2007 10:36:58 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:37:23 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_SH2D1A_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1d1z.
| PDB = {{PDB2|1d1z}}, {{PDB2|1d4t}}, {{PDB2|1d4w}}, {{PDB2|1ka6}}, {{PDB2|1ka7}}, {{PDB2|1m27}}
| Name = SH2 domain protein 1A, Duncan's disease (lymphoproliferative syndrome)
| HGNCid = 10820
| Symbol = SH2D1A
| AltSymbols =; SAP; DSHP; EBVS; IMD5; LYP; MTCP1; XLP; XLPD
| OMIM = 300490
| ECnumber =
| Homologene = 1762
| MGIid = 1328352
| GeneAtlas_image1 = PBB_GE_SH2D1A_210116_at_tn.png
| GeneAtlas_image2 = PBB_GE_SH2D1A_211209_x_at_tn.png
| GeneAtlas_image3 = PBB_GE_SH2D1A_211210_x_at_tn.png
| Function = {{GNF_GO|id=GO:0005070 |text = SH3/SH2 adaptor activity}}
| Component = {{GNF_GO|id=GO:0005737 |text = cytoplasm}}
| Process = {{GNF_GO|id=GO:0006959 |text = humoral immune response}} {{GNF_GO|id=GO:0006968 |text = cellular defense response}} {{GNF_GO|id=GO:0007242 |text = intracellular signaling cascade}} {{GNF_GO|id=GO:0007267 |text = cell-cell signaling}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 4068
| Hs_Ensembl = ENSG00000183918
| Hs_RefseqProtein = NP_002342
| Hs_RefseqmRNA = NM_002351
| Hs_GenLoc_db =
| Hs_GenLoc_chr = X
| Hs_GenLoc_start = 123307875
| Hs_GenLoc_end = 123334686
| Hs_Uniprot = O60880
| Mm_EntrezGene = 20400
| Mm_Ensembl = ENSMUSG00000005696
| Mm_RefseqmRNA = XM_903301
| Mm_RefseqProtein = XP_908394
| Mm_GenLoc_db =
| Mm_GenLoc_chr = X
| Mm_GenLoc_start = 38747189
| Mm_GenLoc_end = 38766724
| Mm_Uniprot = Q544F1
}}
}}
'''SH2 domain protein 1A, Duncan's disease (lymphoproliferative syndrome)''', also known as '''SH2D1A''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Sumegi J, Seemayer TA, Huang D, ''et al.'' |title=A spectrum of mutations in SH2D1A that causes X-linked lymphoproliferative disease and other Epstein-Barr virus-associated illnesses. |journal=Leuk. Lymphoma |volume=43 |issue= 6 |pages= 1189-201 |year= 2003 |pmid= 12152986 |doi= }}
*{{cite journal | author=Engel P, Eck MJ, Terhorst C |title=The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. |journal=Nat. Rev. Immunol. |volume=3 |issue= 10 |pages= 813-21 |year= 2003 |pmid= 14523387 |doi= 10.1038/nri1202 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on SLC19A1... {November 8, 2007 10:44:41 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:46:16 AM PST}
- CREATED: Created new protein page: SLC19A1 {November 8, 2007 10:46:23 AM PST}
- INFO: Beginning work on SLC25A4... {November 8, 2007 10:24:29 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein SLC25A4 image.jpg {November 8, 2007 10:25:09 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:25:28 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_SLC25A4_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1okc.
| PDB = {{PDB2|1okc}}, {{PDB2|2c3e}}
| Name = Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4
| HGNCid = 10990
| Symbol = SLC25A4
| AltSymbols =; ANT; ANT1; PEO2; PEO3; T1
| OMIM = 103220
| ECnumber =
| Homologene = 36058
| MGIid = 1353495
| GeneAtlas_image1 = PBB_GE_SLC25A4_202825_at_tn.png
| Function = {{GNF_GO|id=GO:0005215 |text = transporter activity}} {{GNF_GO|id=GO:0005488 |text = binding}} {{GNF_GO|id=GO:0015207 |text = adenine transmembrane transporter activity}}
| Component = {{GNF_GO|id=GO:0005739 |text = mitochondrion}} {{GNF_GO|id=GO:0005743 |text = mitochondrial inner membrane}} {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}} {{GNF_GO|id=GO:0016021 |text = integral to membrane}}
| Process = {{GNF_GO|id=GO:0000002 |text = mitochondrial genome maintenance}} {{GNF_GO|id=GO:0006091 |text = generation of precursor metabolites and energy}} {{GNF_GO|id=GO:0006810 |text = transport}} {{GNF_GO|id=GO:0006839 |text = mitochondrial transport}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 291
| Hs_Ensembl = ENSG00000151729
| Hs_RefseqProtein = NP_001142
| Hs_RefseqmRNA = NM_001151
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 4
| Hs_GenLoc_start = 186301392
| Hs_GenLoc_end = 186305418
| Hs_Uniprot = P12235
| Mm_EntrezGene = 11739
| Mm_Ensembl = ENSMUSG00000031633
| Mm_RefseqmRNA = XM_134169
| Mm_RefseqProtein = XP_134169
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 8
| Mm_GenLoc_start = 47705991
| Mm_GenLoc_end = 47709847
| Mm_Uniprot = Q8BVI9
}}
}}
'''Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4''', also known as '''SLC25A4''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Vieira HL, Haouzi D, El Hamel C, ''et al.'' |title=Permeabilization of the mitochondrial inner membrane during apoptosis: impact of the adenine nucleotide translocator. |journal=Cell Death Differ. |volume=7 |issue= 12 |pages= 1146-54 |year= 2001 |pmid= 11175251 |doi= 10.1038/sj.cdd.4400778 }}
*{{cite journal | author=Ferri KF, Jacotot E, Blanco J, ''et al.'' |title=Mitochondrial control of cell death induced by HIV-1-encoded proteins. |journal=Ann. N. Y. Acad. Sci. |volume=926 |issue= |pages= 149-64 |year= 2001 |pmid= 11193032 |doi= }}
*{{cite journal | author=Kino T, Pavlakis GN |title=Partner molecules of accessory protein Vpr of the human immunodeficiency virus type 1. |journal=DNA Cell Biol. |volume=23 |issue= 4 |pages= 193-205 |year= 2004 |pmid= 15142377 |doi= 10.1089/104454904773819789 }}
*{{cite journal | author=Andersen JL, Planelles V |title=The role of Vpr in HIV-1 pathogenesis. |journal=Curr. HIV Res. |volume=3 |issue= 1 |pages= 43-51 |year= 2005 |pmid= 15638722 |doi= }}
*{{cite journal | author=Le Rouzic E, Benichou S |title=The Vpr protein from HIV-1: distinct roles along the viral life cycle. |journal=Retrovirology |volume=2 |issue= |pages= 11 |year= 2006 |pmid= 15725353 |doi= 10.1186/1742-4690-2-11 }}
*{{cite journal | author=Zhao RY, Bukrinsky M, Elder RT |title=HIV-1 viral protein R (Vpr) & host cellular responses. |journal=Indian J. Med. Res. |volume=121 |issue= 4 |pages= 270-86 |year= 2005 |pmid= 15817944 |doi= }}
*{{cite journal | author=Muthumani K, Choo AY, Premkumar A, ''et al.'' |title=Human immunodeficiency virus type 1 (HIV-1) Vpr-regulated cell death: insights into mechanism. |journal=Cell Death Differ. |volume=12 Suppl 1 |issue= |pages= 962-70 |year= 2006 |pmid= 15832179 |doi= 10.1038/sj.cdd.4401583 }}
*{{cite journal | author=Li L, Li HS, Pauza CD, ''et al.'' |title=Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions. |journal=Cell Res. |volume=15 |issue= 11-12 |pages= 923-34 |year= 2006 |pmid= 16354571 |doi= 10.1038/sj.cr.7290370 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on SLC26A4... {November 8, 2007 10:41:29 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:42:05 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Solute carrier family 26, member 4
| HGNCid = 8818
| Symbol = SLC26A4
| AltSymbols =; DFNB4; PDS
| OMIM = 605646
| ECnumber =
| Homologene = 20132
| MGIid = 1346029
| GeneAtlas_image1 = PBB_GE_SLC26A4_206529_x_at_tn.png
| Function = {{GNF_GO|id=GO:0005215 |text = transporter activity}} {{GNF_GO|id=GO:0008271 |text = secondary active sulfate transmembrane transporter activity}} {{GNF_GO|id=GO:0015108 |text = chloride transmembrane transporter activity}} {{GNF_GO|id=GO:0015111 |text = iodide transmembrane transporter activity}} {{GNF_GO|id=GO:0031404 |text = chloride ion binding}}
| Component = {{GNF_GO|id=GO:0016020 |text = membrane}} {{GNF_GO|id=GO:0016021 |text = integral to membrane}}
| Process = {{GNF_GO|id=GO:0006810 |text = transport}} {{GNF_GO|id=GO:0007605 |text = sensory perception of sound}} {{GNF_GO|id=GO:0008272 |text = sulfate transport}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 5172
| Hs_Ensembl = ENSG00000091137
| Hs_RefseqProtein = NP_000432
| Hs_RefseqmRNA = NM_000441
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 7
| Hs_GenLoc_start = 107088316
| Hs_GenLoc_end = 107145490
| Hs_Uniprot = O43511
| Mm_EntrezGene = 23985
| Mm_Ensembl = ENSMUSG00000020651
| Mm_RefseqmRNA = NM_011867
| Mm_RefseqProtein = NP_035997
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 12
| Mm_GenLoc_start = 32101856
| Mm_GenLoc_end = 32145728
| Mm_Uniprot = Q9R155
}}
}}
'''Solute carrier family 26, member 4''', also known as '''SLC26A4''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Mutations in this gene are associated with Pendred syndrome, the most common form of syndromic deafness, an autosomal-recessive disease. It is highly homologous to the SLC26A3 gene; they have similar genomic structures and this gene is located 3' of the SLC26A3 gene. The encoded protein has homology to sulfate transporters.<ref>{{cite web | title = Entrez Gene: SLC26A4 solute carrier family 26, member 4| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5172| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Markovich D |title=Physiological roles and regulation of mammalian sulfate transporters. |journal=Physiol. Rev. |volume=81 |issue= 4 |pages= 1499-533 |year= 2001 |pmid= 11581495 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on SOD3... {November 8, 2007 10:46:23 AM PST}
- CREATE: Found no pages, creating new page. {November 8, 2007 10:46:57 AM PST}
- CREATED: Created new protein page: SOD3 {November 8, 2007 10:47:04 AM PST}
- INFO: Beginning work on VIP... {November 8, 2007 10:47:04 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:47:49 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Vasoactive intestinal peptide
| HGNCid = 12693
| Symbol = VIP
| AltSymbols =; MGC13587; PHM27
| OMIM = 192320
| ECnumber =
| Homologene = 2539
| MGIid = 98933
| GeneAtlas_image1 = PBB_GE_VIP_206577_at_tn.png
| Function = {{GNF_GO|id=GO:0005179 |text = hormone activity}} {{GNF_GO|id=GO:0005184 |text = neuropeptide hormone activity}}
| Component = {{GNF_GO|id=GO:0005576 |text = extracellular region}} {{GNF_GO|id=GO:0005625 |text = soluble fraction}}
| Process = {{GNF_GO|id=GO:0006936 |text = muscle contraction}} {{GNF_GO|id=GO:0007186 |text = G-protein coupled receptor protein signaling pathway}} {{GNF_GO|id=GO:0007267 |text = cell-cell signaling}} {{GNF_GO|id=GO:0007589 |text = fluid secretion}} {{GNF_GO|id=GO:0008284 |text = positive regulation of cell proliferation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 7432
| Hs_Ensembl = ENSG00000146469
| Hs_RefseqProtein = NP_003372
| Hs_RefseqmRNA = NM_003381
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 6
| Hs_GenLoc_start = 153113626
| Hs_GenLoc_end = 153122593
| Hs_Uniprot = P01282
| Mm_EntrezGene = 22353
| Mm_Ensembl = ENSMUSG00000019772
| Mm_RefseqmRNA = NM_011702
| Mm_RefseqProtein = NP_035832
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 10
| Mm_GenLoc_start = 4698927
| Mm_GenLoc_end = 4707323
| Mm_Uniprot = P32648
}}
}}
'''Vasoactive intestinal peptide''', also known as '''VIP''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = The protein encoded by this gene belongs to the glucagon family. It stimulates myocardial contractility, causes vasodilation, increases glycogenolysis, lowers arterial blood pressure and relaxes the smooth muscle of trachea, stomach and gall bladder. Alternative splicing occurs at this locus and two transcript variants encoding distinct isoforms have been identified.<ref>{{cite web | title = Entrez Gene: VIP vasoactive intestinal peptide| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7432| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Fahrenkrug J |title=Gut/brain peptides in the genital tract: VIP and PACAP. |journal=Scand. J. Clin. Lab. Invest. Suppl. |volume=234 |issue= |pages= 35-9 |year= 2002 |pmid= 11713978 |doi= }}
*{{cite journal | author=Delgado M, Pozo D, Ganea D |title=The significance of vasoactive intestinal peptide in immunomodulation. |journal=Pharmacol. Rev. |volume=56 |issue= 2 |pages= 249-90 |year= 2004 |pmid= 15169929 |doi= 10.1124/pr.56.2.7 }}
*{{cite journal | author=Conconi MT, Spinazzi R, Nussdorfer GG |title=Endogenous ligands of PACAP/VIP receptors in the autocrine-paracrine regulation of the adrenal gland. |journal=Int. Rev. Cytol. |volume=249 |issue= |pages= 1-51 |year= 2006 |pmid= 16697281 |doi= 10.1016/S0074-7696(06)49001-X }}
*{{cite journal | author=Hill JM |title=Vasoactive intestinal peptide in neurodevelopmental disorders: therapeutic potential. |journal=Curr. Pharm. Des. |volume=13 |issue= 11 |pages= 1079-89 |year= 2007 |pmid= 17430171 |doi= }}
*{{cite journal | author=Gonzalez-Rey E, Varela N, Chorny A, Delgado M |title=Therapeutical approaches of vasoactive intestinal peptide as a pleiotropic immunomodulator. |journal=Curr. Pharm. Des. |volume=13 |issue= 11 |pages= 1113-39 |year= 2007 |pmid= 17430175 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on VIPR1... {November 8, 2007 10:47:49 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 8, 2007 10:48:54 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Vasoactive intestinal peptide receptor 1
| HGNCid = 12694
| Symbol = VIPR1
| AltSymbols =; II; FLJ41949; HVR1; PACAP-R-2; RDC1; VAPC1; VIPR; VIRG; VPAC1
| OMIM = 192321
| ECnumber =
| Homologene = 3399
| MGIid = 109272
| GeneAtlas_image1 = PBB_GE_VIPR1_205019_s_at_tn.png
| Function = {{GNF_GO|id=GO:0004872 |text = receptor activity}} {{GNF_GO|id=GO:0004930 |text = G-protein coupled receptor activity}} {{GNF_GO|id=GO:0004999 |text = vasoactive intestinal polypeptide receptor activity}}
| Component = {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}}
| Process = {{GNF_GO|id=GO:0006936 |text = muscle contraction}} {{GNF_GO|id=GO:0006955 |text = immune response}} {{GNF_GO|id=GO:0007165 |text = signal transduction}} {{GNF_GO|id=GO:0007186 |text = G-protein coupled receptor protein signaling pathway}} {{GNF_GO|id=GO:0007187 |text = G-protein signaling, coupled to cyclic nucleotide second messenger}} {{GNF_GO|id=GO:0007268 |text = synaptic transmission}} {{GNF_GO|id=GO:0007586 |text = digestion}} {{GNF_GO|id=GO:0008284 |text = positive regulation of cell proliferation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 7433
| Hs_Ensembl = ENSG00000114812
| Hs_RefseqProtein = NP_004615
| Hs_RefseqmRNA = NM_004624
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 3
| Hs_GenLoc_start = 42519121
| Hs_GenLoc_end = 42554064
| Hs_Uniprot = P32241
| Mm_EntrezGene = 22354
| Mm_Ensembl = ENSMUSG00000032528
| Mm_RefseqmRNA = NM_011703
| Mm_RefseqProtein = NP_035833
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 9
| Mm_GenLoc_start = 121491414
| Mm_GenLoc_end = 121519332
| Mm_Uniprot = P97751
}}
}}
'''Vasoactive intestinal peptide receptor 1''', also known as '''VIPR1''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a receptor for vasoactive intestinal peptide, a small neuropeptide. Vasoactive intestinal peptide is involved in smooth muscle relaxation, exocrine and endocrine secretion, and water and ion flux in lung and intestinal epithelia. Its actions are effected through integral membrane receptors associated with a guanine nucleotide binding protein which activates adenylate cyclase.<ref>{{cite web | title = Entrez Gene: VIPR1 vasoactive intestinal peptide receptor 1| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7433| accessdate = }}</ref>
}}
==References==
{{reflist}}
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
}}
{{refend}}
{{protein-stub}}
end log.
 {November 8, 2007 10:39:55 AM PST}
{November 8, 2007 10:39:55 AM PST} 
