User:JeanOhm/golgi
This is a practice page for Golgi apparatus improvements

The Golgi apparatus (/ˈɡoʊldʒiː/), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in eukaryotic cells. It was reported in 1898[1] by the Italian scientist Camillo Golgi and first named after him in 1910.[2][3][a] Various aspects of the Golgi have been reviewed.[5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22]
Part of the cellular endomembrane system, the Golgi apparatus is best known for packaging proteins into membrane-bound vesicles inside the cell before the vesicles are sent to their destination. It is of particular importance in processing proteins for secretion, containing a set of glycosylation enzymes that attach various sugar monomers to proteins as the proteins move through the apparatus. The Golgi apparatus resides at the intersection of the secretory, lysosomal, and endocytic pathways. It is also involved in cell cycle control and amino acid sensing. The Golgi is a dynamic structure in living cells and is highly variable in structure in different organisms.


Discovery[edit]

Owing to its large size and distinctive structure, the Golgi apparatus was one of the first organelles to be discovered and observed in detail. It was discovered in 1898 by Italian physician Camillo Golgi during an investigation of the nervous system.[1] After first observing it under his microscope, he termed the structure the "internal reticular apparatus". Early references to the Golgi referred to it by various names including the "Golgi–Holmgren apparatus", "Golgi–Holmgren ducts", and "Golgi–Kopsch apparatus".[3] Some doubted the discovery at first, arguing that the appearance of the structure was merely an optical illusion created by the observation technique used by Golgi.[3] With the development of modern microscopes in the 20th century, the discovery was confirmed.[24][b]
Structure[edit]
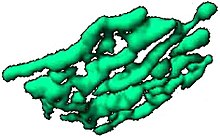



The Golgi apparatus is often composed of flattened lipid bilayer membrane-enclosed disks known as cisternae (singular: cisterna) which originate from vesicles that bud off of "smooth" (ribosome-free) regions of the rough endoplasmic reticulum (RER). Often 3 to 5 cisternae are present in a single Golgi stack, but many more can be present.[25][26] The cis aspect of the stack is defined as that closest to the RER, while the trans aspect is on the opposite face of the stack. The cisternae have sometimes been reported to be connected by tubules.[27][28][29] The Trans Golgi Network (TGN) is often found after the trans face of the stack.[21] In vertebrates a structure known as the Endoplasmic Reticulum-Golgi Intermediate Complex (ERGIC) is observed between the RER and the Golgi stack.[21]
There are structural and organizational differences in the Golgi apparatus among eukaryotes. In some yeasts, Golgi stacking is not observed. The yeast Pichia pastoris does have stacked Golgi, while the baker's (and brewer's) yeast Saccharomyces cerevisiae normally have their cisternae dispersed in the cytoplasm,[11]. As mentioned in the legend of a movie on this page, the names early and late Golgi are used in Saccharomyces cerevisiae, and those names roughly correspond to cis and trans, respectively.
In the intracellular parasite of animals, microsporidia, there is an interlaced network of tubules (MIN) that functions like the Golgi.[30][c] In contrast, Ostreococcus tauri (an alga which is the smallest known free-living eukaryote) has a single well-defined Golgi stack. In vertebrates, as shown by images on this page, Golgi stacks are sometimes connected by tubules to form ribbons.[6] Golgi matrix proteins are important for the structure of Golgi components.[6][7][31][32] In plants, individual Golgi stacks have been seen to be connected by tubules, although not as a ribbon.[5]
In summary, the ancestral Golgi structure appears to have been a stack, with several independent losses of stacking in various lineages[11] and the development of Golgi ribbons in vertebrates during evolution.[6]

Subcellular localization[edit]


Among eukaryotes, the subcellular localization of the Golgi apparatus differs. In mammals, a single Golgi ribbon is usually located near the cell nucleus, close to the centrosome[15]. This is shown especially well in mammalian cells in culture, which have their endoplasmic reticulum spread out over the extent of the cytoplasm, while the Golgi remains near the nucleus.[13] Localization and tubular connections of the Golgi apparatus are dependent on microtubules.[16] In mammalian cells engineered to express a particular Golgi matrix protein, GMAP210,[d] on their mitochondria, their mitochondria become clustered near the centrosome.[33] If microtubules are experimentally depolymerized, then the Golgi apparatus loses connections and becomes individual stacks throughout the cytoplasm.[34] In baker's yeast, Golgi cisternae are scattered throughout the cytoplasm.[23] In plants, Golgi stacks are not concentrated at the centrosomal region, do not form Golgi ribbons[5] and move at several micrometers per second in relation to the ER.[17] Until recently, organization of the plant Golgi was thought to depend on actin cables and not microtubules,[5] but now it is believed that microtubules are involved in "fine-tuning" plant Golgi.[14] A common, but not universal, feature among Golgi is that they are adjacent to RER exit sites.[11]
Biochemical functions in the lumen[edit]


The best-known biochemical function of the Golgi is the continuation of sugar modifications to glycoproteins and glycolipids (collectively, the glycome) that was initiated in the endoplasmic reticulum.[19] Glycosyltransferases[e] are the enzymes responsible for building up larger sugar chains while glycoside hydrolase enzymes break down the bonds between the sugar residues. The production of the final product is not uniformly in the direction of increasing length or complexity of the glycan, as exemplified in the image. In cells with defined stacks, individual cisternae or groups of cisternae have different assortments of enzymes, allowing for progressive processing of cargo molecules as they travel from the cis to the trans Golgi face.[34][35] That is true in plants, which also synthesize the complex polysaccharides of their cell walls in the Golgi lumen using enzymes localized to different parts of the Golgi.[36]
There are a variety of other biochemical alterations in the Golgi. Sulfation of tyrosines and carbohydrates occurs within the TGN.[37][34] Phosphorylation is important in many cellular processes, and occurs in various locations, including the Golgi,[38][34], which also contains phosphatases that can remove phosphates.[39] Lipidation reactions such as palmitoylation, which covalently attach a lipid residue to a protein, also occur in the Golgi.[40][41] Recently, a protease in the Golgi that cleaves multiple proteins has been reported to be important biochemically,[42] in cell biology[43] and virology.[44]
Cargo trafficking and sorting[edit]

There are many models for how molecules are transported from the rough endoplsmic reticulum to their final destinations, but none of the models can explain all experimental observations.[8] Most (but not all[45]) biologists agree, however, that vesicular transport is involved.

Vesicular transport[edit]

Animation showing the process


The 2013 Nobel Prize in Physiology or Medicine was awarded for "discoveries of machinery regulating vesicle traffic, a major transport system in our cells"[47][48][f] There is obviously a vast amount known about the trafficking of proteins made in the RER. The process starts with the production of a COPII vesicle from an endoplasmic reticulum exit site. The COPII vesicle then moves toward the Golgi, in some instances fusing first with the ERGIC. There is at least one report that COPII vesicles can also be generated from the ERGIC to transport materials to other parts of the cell.[49] A special case is the transport of collagen,[g] which is much larger than standard COPII vesicle and is instead packaged into "mega carriers".[18][50] [51]
The directed movement of vesicles can be accomplished by tethering them to molecular motors that move along microtubules as shown in the accompanying figure.[52] The motors only move in one direction on the micotubule, but there are different motors for the "forward" and "reverse" directions.
It should be noted that not all important cargo molecules are necessarily incorporated into vesicles that migrate away from the ER. There are close contact sites that have been reported to allow lipid transfer between the ER and Golgi,[53] Close contacts have been incorporated into a "hug and kiss" model[54] for the transfer of cargo from the ER to Golgi.[55]
The COPII vesicles carry materials that should remain part of the ER, as well as molecules destined for the Golgi and beyond. In order to return the ER components to the ER from the Golgi or ERGIC, COPI vesicles are formed for the retrograde transport. If that retrograde transport is inhibited, the Golgi actually swells in all dimensions.[56] The mechanism by which the vesicles and other organelles fuse involves SNARE proteins and is explained in the SNARE article and lipid bilayer fusion.
In fairness, as of 2015, four expert botanists could not reach a consensus about transport from the ER to Golgi in higher plants.[17]
Movement between/of the cisternae[edit]


Again, there are many models of how molecules transit from the cis to trans face,[8] not one of which is consistent with all currently available data in all species. One of the most popular early models was that of cisternal maturation, in which cisternae at the cis face would gradually mature while migrating towards the trans face, and would be replaced by newly formed cisternae produced by maturation of the ERGIC (or similar structures in plants).[26] That model requires transport of molecules that are known to be resident in only one part of the Golgi in a retrograde direction from more trans to more cis cisternae as the cisternae mature. The reports of transport of cargoes through Golgi cisternae that are "stapled"[57] or "glued"[58] argue against this model.
Another model theorizes that the Golgi cisternae stay in place, while secretory cargo molecules are somehow transported toward the trans face, either in vesicles or tubules or through direct connections. Images and videos in this section show connections at the edges of stacks. These types of connections, as well as the finding that differently-sized cargo molecules move through the Golgi at different rates, has led to the idea that transport of soluble cargo occurs by diffussion through those connections.[46]

Another popular model is that of anterograde (cis to trans) movement of cargo in vesicles. Vesicles have been reported to form from, and "percolate" between, cisternae in a stack[35][23][h] and even between cisternae of different stacks.[59]
As of 2016 there is still "heated" debate among experts about how cargo is trafficked through the Golgi and evidence in support of the the various hypotheses is "mixed at best".[60][61] The authors of a 2017 review wrote that nothing about the Golgi "can be stated with absolute clarity".[22]
Exit to other sites[edit]


The accompanying figure shows complicated trafficking pathways "downstream" from the Golgi. One complication involves the "exact" identity of the structures labeled 1 and 2. Different authors attached different names to some of them[i] but in this article both will be described collectively as simply "trans Golgi".
The selection/sorting and packaging of cargo involve the vesicular transport Adaptor Protein complexes AP-1 through 5, retromer, exomer and other cargo adaptors. In order to avoid a significant redundancy with the other articles, only selected aspects of cargo exit will be presented.
One well studied example of specificity of sorting and exit to specific sites involves delivery of cargo molecules to the basolateral membrane of polarized epithelial cells.[65] The selection of cargo molecules by the AP complexes involves interactions of specific AP complex subunits with specific amino acid motifs on proteins. The AP1 complex has a medium sized (mu, μ or M) subunit, but in humans there are 2 genes encoding slightly different proteins, AP1M1 and AP1M2. AP1M1 is expressed in all cells, while AP1M2 is expressed in only some cells, and the protein product of AP1M2 binds better than AP1M1 protein to basolateral signal motifs of basolateral cargo proteins.
In contrast to those specific protein-protein interactions, there is evidence that specific protein clustering into "lipid rafts" and the incorporation of those rafts into vesicles that fuse with the apical plasma membrane is important for delivery of cargo to the apical surface.[65][66] While important signalling sites for a variety of apical proteins have been reported, they can be anywhere on the protein, not just residing inside the lipid bilayer.[65]
Note that the trans Golgi can be involved in a circulation of cargo to the plasma membrane and back to the Golgi through endosomal intermediates. Cargo that needs to be degraded can be shunted away to the lysosome. The diagram may give the impression that the anterograde transport is only via vesicles, but the accompanying video clearly shows tubular transport,[j] and as of 2015 "it is becoming increasingly accepted that together with vesicles, tubules play a major role in the transfer of cargo between different cellular compartments."[67]


In some cases, the Golgi is not even involved in transport of cargo to the plasma membrane. For example, in the stem cell shown in the accompanying figure, endoplasmic reticulum is located throughout the cell, while Golgi are absent in the basal region. Proteins still get to the basal plasma membrane, although they do not have the posttranslational modifications, indicating that there can be direct transfer from ER to the plasma membrane.[68]
The signal that targets proteins from the trans Golgi to the lysosome is not even protein, rather it is a mannose-6-phosphate on the glycan chain.[34]
The accompanying diagram indicates that COPI vesicles transport ER resident materials back to the ER. In fact, numerous studies have shown that all Golgi components continuously recycle back to the ER.[69]
cell cycle and growth control[edit]

In addition to other cell cycle checkpoints, the Golgi organization also provides a mitotic checkpoint. When the "unlinking" of the stacks in a Golgi ribbon is inhibited, cell cycle progression is blocked.[9]

The unlinking is caused by phosphorylation of the Golgi matrix protein GRASP65 at residue Ser277 by one of the c-Jun N-terminal kinases, JNK2.[71]
After unlinking, the Golgi stacks become unstacked and then form vesicular or tubular fragments that have been described as a “haze”.[9] After mitosis, the Golgi stacks must be reformed. That reassembly is dependent on the GRASP proteins, both in vitro and in vivo, as shown in the accompanying image.
There is evidence of a Golgi-localized type of mTORC1, which may be part of an amino acid starvation signaling hub located on the Golgi and involved in cell growth control.[72]
Relationships with cytoskeleton/other organelles[edit]


The Golgi matrix protein GMAP210 has been shown to co-purify with microtubules, and changes in the expression of GMAP210 perturbs not only Golgi, but also microtubule network organization. GMAP210 associates with the γ-tubulin small complex, which is required for nucleation of microtubules.[73] The Golgi is known to be a microtubule organizing center, and more than one protein is involved.[10][12]

Membrane contact sites (MCSs) are sites where two membranes come near to each other, usually less than 30 nm apart.[75] MCSs have been observed between the Golgi and the phagophore, the sack-like structure involved in autophagosome formation. Approximately

more details of VAPs[76]
Lipid exchange schematic[76]
20% of phagophores at any one time are closely associated with Golgi membranes, which may be a source of lipids for the expanding phagophore.[74] There is a report that Golgi cisternae can directly develop into autophagosomes.[77]
The Golgi "interactome" has been reported to also include the ER, mitochondria, lysosomes, lipid droplets and peroxisomes."[78] The best characterized are those between the Golgi and ER.

MCSs are held together by protein tethers,rather than ctoskeletal elements.[75]. CASP[k] has been reported to be such a tether for the ER-Golgi MCSs.[17] There are several other proteins that function in the ER-Golgi MCSs. VAPs A and B are in the ER membrane and bind to CERT, FAPP2, NIR2 and OSBP which associate with the Golgi through their Pleckstrin homology domains recognizing phosphatidylinositol 4-phosphate.[20] The 4 proteins all contain FFAT motifs that interacts with the VAPs.[20] These proteins are involved in the transfer of lipids from one membrane to the other.[20] For example, there is a gradient of cholesterol in mammalian cells, with higher concentration in the plasma membrane and lower in the ER membranes, and the gradient is produced by the exchange of cholesterol for phosphatidylinositol 4-phosphate.[80]
The toxin ricin travels from the plasma membrane through the Golgi and to the ER via a retrograde movement without entering the cytoplasm.[79] This pathway is thought to use the same mechanism as in Endoplasmic-reticulum-associated protein degradation (ERAD) of damaged proteins from the Golgi.[79]

Review article model[8]
Rapid partitioning in a 2 phase model[edit]
A very different model of Golgi functioning has been proposed,[81] and explains some aspects that cannot be explained by "cisernal maturation".[8] In this model, the Golgi has 2 "phases" or "domains" (left vs right in both external images), and cargo would exit stochastically from the domains at any level (cis, medial or trans) to their next destination.
Golgipathies[edit]

and human from mutation in GMAP-210[82]
The term "Golgipathies" was coined in a review of Cohen syndrome, progressive cerebello-cerebral atrophy type 2 PCCA2 syndrome (mutation in VPS53), Warburg-micro syndrome, MRT13 (mutation in TRAPPC9), A neuromuscular syndrome (mutation in GOLGA2), Dyggve-Melchior-Clausen syndrome (mutation in DYM) and Congenital disorders of glycosylation (mutations in COG1, COG2, COG7 or COG8)[83] Many other disease associated with post-translational modifications in the Golgi have also been reviewed.[84]
Mice engineered to lack the golgin encoded by the USO1 where found to suffer early embryonic death.[85] A less severe, but still lethal neonatally, skeletal dysplasia was found in mice lacking GMAP-210, and human achondrogenesis type 1A is associated with homzygous loss of function mutations in the corresponding human gene.[82]
Gallery[edit]

Neonatal mice littermates heterozygous or homozygous for a GMAP210 mutation.[86]

The F-box protein FBXW8 is required for normal Golgi (highlighted yellow) ribbon formation in rat neurons.[87]
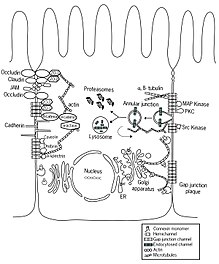
The trans Golgi must send cargo to the correct locations in polarized cells, such as in cells of epithelia[88]

Atomic-force microscopic image of part of a Golgi apparatus isolated from HeLa cells.[89]

The vesicle tether GMAP210 is thought to have a hinge that allows the vesicle to approach the Golgi, while GCC185 is thought to be more flexible and collapse onto the Golgi.[90]

Arabidopsis COG3 and COG8 are required for normal Golgi structure.[91]

ELKS interacts with RAB6 to direct cargo vesicles along microtubules to melanosomes. The flourescent image is from the movie, which shows bright green vesicles moving in a human melanocyte cell line. The bottom panel shows microtubules in red, mobile vesicles in bright green, and melanosomes in the duller green.[92]
Notes[edit]
- ^ An individual Golgi stack is sometimes called a dictyosome (from Greek dictyon: net + soma: body[4]
- ^ Some illustrations from the 1920's are reproduced in the previous reference by Goldfischer
- ^ Note the similarity between the Microsporidia Interlaced Network (in brown) and the mammalian Golgi shown here
- ^ "GMAP" is derived from Golgi Microtubule Associated Protein. It is known to bind to gamma tubulin.
- ^ There are more than 250 different glycosyltransferase enzymes in the Golgi.[22]
- ^ Dr. Randy Schekman's Nobel lecture cited in the previous reference recounts many topics relevant to this article.
- ^ Collagen accounts for approximately 25% of the dry weight of proteins present in the readers of this article, so it is an important special case!
- ^ To illustrate how uncertain the field is, the publications from the same lab write about percolating vesicles and cisternal maturation!
- ^ Some describe 1 as TGN and 2 as some sort of endosomal or tubular endosomal cargo recycling compartment, but even then will write "1/2" or speak "trans Golgi" to describe both. One publication defined a "nascent vesicle site" that "was adjacent to, if not a sub-compartment of, the trans-Golgi network".[63] Considering the variety of Golgi structures presented in this article and the variabilty of the TGN analyzed in this research paper,[64] please excuse the simplified nomenclature.
- ^ Note that by confocal light microscopy the Golgi is not as precisely visualized as in the electron micrographs. It can only be seen as a "blob".
- ^ CASP is an Alternatively Spliced Product from the CUX1 gene.
References[edit]
- ^ a b Golgi, C. (1898). Intorno alla struttura delle cellule nervose. Bollettino della Società Medico-Chirurgica di Pavia 13(1): 316.
- ^ Besta, C (1910). "Sull'apparato reticolare interno (apparato di Golgi) della cellula nervosa". Anatomischer Anzeiger. 36: 476–486.
- ^ a b c Fabene PF, Bentivoglio M (1998). "1898–1998: Camillo Golgi and "the Golgi": one hundred years of terminological clones". Brain Res. Bull. 47 (3): 195–8. doi:10.1016/S0361-9230(98)00079-3. PMID 9865849.
- ^ anon. "dictyosome". Dictionary.com. Dictionary.com, LLC. Retrieved 2 April 2017.
- ^ a b c d Nakano A, Luini A (2010). "Passage through the Golgi". Curr Opin Cell Biol. 22 (4): 471–8. doi:10.1016/j.ceb.2010.05.003. PMID 20605430.
- ^ a b c d Wei JH, Seemann J. (2010). "Unraveling the Golgi ribbon". Traffic. 11 (11): 1391–400. doi:10.1111/j.1600-0854.2010.01114.x. PMID 21040294.
- ^ a b Lowe, M (2011). "Structural organization of the Golgi apparatus". Current opinion in cell biology. 23 (1): 85–93. doi:10.1016/j.ceb.2010.10.004. PMID 21071196.
- ^ a b c d e Glick BS, Luini A (2011). "Models for Golgi traffic: a critical assessment". Cold Spring Harb Perspect Biol. 3 (11): a005215. doi:10.1101/cshperspect.a005215. PMID 21875986.
- ^ a b c Corda D, Barretta ML, Cervigni RI, Colanzi A (2012). "Golgi complex fragmentation in G2/M transition: An organelle-based cell-cycle checkpoint". IUBMB Life. 64 (8): 661–70. doi:10.1002/iub.1054. PMID 22730233.
- ^ a b c Millarte V, Farhan H (2012). "The Golgi in cell migration: regulation by signal transduction and its implications for cancer cell metastasis". TheScientificWorldJournal. 2012: 498278. doi:10.1100/2012/498278. PMC 3353474. PMID 22623902.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d Suda, Y; Nakano A, A (2012). "The yeast Golgi apparatus". Traffic. 13 (4): 505–10. doi:10.1111/j.1600-0854.2011.01316.x. PMID 22132734.
- ^ a b c Zhu X, Kaverina I (2013). "Golgi as an MTOC: making microtubules for its own good". Histochemistry and Cell Biology. 140 (3): 361–7. doi:10.1007/s00418-013-1119-4. PMC 3748218. PMID 23821162.
- ^ a b Brandizzi, F and Barlowe, C. "Organization of the ER–Golgi interface for membrane traffic control". Nat Rev Mol Cell Biol. 2013 Jun; 14(6): 382–392. National Center for Biotechnology Information,. Retrieved 3 April 2017.
{{cite web}}: CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link) - ^ a b Brandizzi F, Wasteneys GO., Plant J. 2013 Jul;75(2):339-49. doi: 10.1111/tpj.12227. "Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules". onlinelibrary.wiley.com. John Wiley & Sons, Inc. Retrieved 12 April 2017.
{{cite web}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ a b Rios, RM, Philosophical Transactions of the Royal Society of London - Series B: Biological Sciences. 369(1650), 2014 Sep 05. "The centrosome–Golgi apparatus nexus". The Royal Society Publishing. Retrieved 4 April 2017.
{{cite web}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ a b Gurel PS, Hatch AL, Higgs HN (2014). "Connecting the cytoskeleton to the endoplasmic reticulum and Golgi". Current Biology : CB. 24 (14): R660–72. doi:10.1016/j.cub.2014.05.033. PMC 4174561. PMID 25050967.
- ^ a b c d Robinson DG, Brandizzi F, Hawes C, Nakano A, Plant Physiol. 2015 Jun;168(2):393-406. doi: 10.1104/pp.15.00124. "Vesicles versus Tubes: Is Endoplasmic Reticulum-Golgi Transport in Plants Fundamentally Different from Other Eukaryotes?". www.ncbi.nlm.nih.gov. American Society of Plant Biologists. Retrieved 11 April 2017.
{{cite web}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ a b Malhotra V, Erlmann P. (2015). "The pathway of collagen secretion". Annu Rev Cell Dev Biol. 31: 109–24. doi:10.1146/annurev-cellbio-100913-013002. PMID 26422332.
- ^ a b Kellokumpu S, Hassinen A, Glumoff T (2016). "Glycosyltransferase complexes in eukaryotes: long-known, prevalent but still unrecognized". Cell Mol Life Sci. 73 (2): 305–25. doi:10.1007/s00018-015-2066-0. PMID 26474840.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e Phillips MJ, Voeltz GK (2016). "Structure and function of ER membrane contact sites with other organelles". Nature Reviews. Molecular Cell Biology. 17 (2): 69–82. doi:10.1038/nrm.2015.8. PMC 5117888. PMID 26627931.
- ^ a b c Luini A, Parashuraman S (2016). "Signaling at the Golgi: sensing and controlling the membrane fluxes". Curr Opin Cell Biol. 39: 37-42. doi:10.1016/j.ceb.2016.01.014. PMID 26908115.
- ^ a b c Mironov AA, Sesorova IS, Seliverstova EV, Beznoussenko GV (2017). "Different Golgi ultrastructure across species and tissues: Implications under functional and pathological conditions, and an attempt at classification". Tissue Cell. 49 (2 Pt A): 186-201. doi:10.1016/j.tice.2016.12.002. PMID 28007425.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Papanikou E, Day KJ, Austin J, Glick BS (2015). "COPI selectively drives maturation of the early Golgi". eLife. 4. doi:10.7554/eLife.13232. PMC 4758959. PMID 26709839.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Cite error: The named reference "pmid26709839" was defined multiple times with different content (see the help page). - ^ Goldfischer, S. "The internal reticular apparatus of Camillo Golgi: a complex, heterogeneous organelle, enriched in acid, neutral, and alkaline phosphatases, and involved in glycosylation, secretion, membrane flow, lysosome formation, and intracellular digestion". J Histochem Cytochem. 1982 Jul;30(7):717-33. DOI: 10.1177/30.7.6286754. Retrieved 1 April 2017.
- ^ Donohoe, B; et al. "Electron micrographs of a Golgi stack and Golgi cisternae of high-pressure frozen and freeze-substituted cells of the scale-forming alga Scherffelia dubia". onlinelibrary.wiley.com/. John Wiley & Sons, Inc. Retrieved 3 June 2017.
{{cite web}}: Explicit use of et al. in:|first1=(help) - ^ a b Donohoe BS, Kang BH, Gerl MJ, Gergely ZR, McMichael CM, Bednarek SY, Staehelin LA., Traffic. 2013 May;14(5):551-67. doi: 10.1111/tra.12052. "Cis-Golgi cisternal assembly and biosynthetic activation occur sequentially in plants and algae". onlinelibrary.wiley.com. John Wiley & Sons, Inc. Retrieved 12 April 2017.
{{cite web}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer SR, Rabouille C, Rothman JE, Warren G, Wieland FT. (2009). "Journeys through the Golgi--taking stock in a new era". J Cell Biol. 187 (4): 449–53. doi:10.1083/jcb.200909011. PMID 19948493.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c San Pietro E, Capestrano M, Polishchuk EV, DiPentima A, Trucco A, Zizza P, Mariggiò S, Pulvirenti T, Sallese M, Tete S, Mironov AA, Leslie CC, Corda D, Luini A, Polishchuk RS. (2009). "Group IV phospholipase A(2)alpha controls the formation of inter-cisternal continuities involved in intra-Golgi transport". PLoS Biol. 7: e1000194. doi:10.1371/journal.pbio.1000194. PMID 19753100.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Park SY, Yang JS, Schmider AB, Soberman RJ, Hsu VW (2015). "Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42". Nature. 521 (7553): 529–32. doi:10.1038/nature14457. PMID 25945738.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Beznoussenko GV, Dolgikh VV, Seliverstova EV, Semenov PB, Tokarev YS, Trucco A, Micaroni M, Di Giandomenico D, Auinger P, Senderskiy IV, Skarlato SO, Snigirevskaya ES, Komissarchik YY, Pavelka M, De Matteis MA, Luini A, Sokolova YY, Mironov AA., J Cell Sci. 2007 Apr 1;120(Pt 7):1288-98. "Analogs of the Golgi complex in microsporidia: structure and avesicular mechanisms of function". Journal of Cell Science. Retrieved 3 April 2017.
{{cite web}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Xiang Y, Wang Y. (2011). "New components of the Golgi matrix". Cell Tissue Res. 344 (3): 365-79. doi:10.1007/s00441-011-1166-x. PMID 21494806.
- ^ Osterrieder, A (2012). "Tales of tethers and tentacles: golgins in plants". J Microsc. 247 (1): 68-77. doi:10.1111/j.1365-2818.2012.03620.x. PMID 22591132.
- ^ Ríos RM, Sanchís A, Tassin AM, Fedriani C, Bornens M (2004). "GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation". Cell. 118 (3): 323–35. doi:10.1016/j.cell.2004.07.012. PMID 15294158.
- ^ a b c d e Alberts, Bruce; et al. Molecular Biology of the Cell. Garland Publishing. ISBN 978-0-8153-1619-0.
- ^ a b Day KJ, Staehelin LA, Glick BS (2013). "A three-stage model of Golgi structure and function". Histochemistry and Cell Biology. 140 (3): 239–49. doi:10.1007/s00418-013-1128-3. PMC 3779436. PMID 23881164.
- ^ Zhang GF, Staehelin LA., Plant Physiol. 1992 Jul;99(3):1070-83. PMC 1080586. "Functional compartmentation of the Golgi apparatus of plant cells : immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells" (PDF). ncbi.nlm.nih.gov. American Society of Plant Biologists. Retrieved 12 April 2017.
{{cite web}}: External link in|first1= - ^ Hartmann-Fatu C, Trusch F, Moll CN, Michin I, Hassinen A, Kellokumpu S, Bayer P (2015). "Heterodimers of tyrosylprotein sulfotransferases suggest existence of a higher organization level of transferases in the membrane of the trans-Golgi apparatus". J Mol Biol. 427: 1404–12. doi:10.1016/j.jmb.2015.01.021. PMID 25660941.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sreelatha A, Kinch LN, Tagliabracci VS (2015). "The secretory pathway kinases". Biochimica Et Biophysica Acta. 1854 (10 Pt B): 1687–93. doi:10.1016/j.bbapap.2015.03.015. PMC 4577305. PMID 25862977.
- ^ Tang X, Benesch MG, Brindley DN (2015). "Lipid phosphate phosphatases and their roles in mammalian physiology and pathology". Journal of Lipid Research. 56 (11): 2048–60. doi:10.1194/jlr.R058362. PMC 4617392. PMID 25814022.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Hesse D, Radloff K, Jaschke A, Lagerpusch M, Chung B, Tailleux A, Staels B, Schürmann A (2014). "Hepatic trans-Golgi action coordinated by the GTPase ARFRP1 is crucial for lipoprotein lipidation and assembly". Journal of Lipid Research. 55 (1): 41–52. doi:10.1194/jlr.M040089. PMC 3927470. PMID 24186947.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Smagris E, Gilyard S, BasuRay S, Cohen JC, Hobbs HH (2016). "Inactivation of Tm6sf2, a Gene Defective in Fatty Liver Disease, Impairs Lipidation but Not Secretion of Very Low Density Lipoproteins". The Journal of Biological Chemistry. 291 (20): 10659–76. doi:10.1074/jbc.M116.719955. PMC 4865914. PMID 27013658.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Kuhn PH; Voss M; Haug-Kroper M; Schroder B; Schepers U; Brase S; Haass C; Lichtenthaler SF; Fluhrer R (2015). "Secretome analysis identifies novel signal Peptide peptidase-like 3 (Sppl3) substrates and reveals a role of Sppl3 in multiple Golgi glycosylation pathways". Molecular & Cellular Proteomics. 14 (6): 1584–98. doi:10.1074/mcp.M115.048298. PMID 25827571.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Hamblet CE; Makowski SL; Tritapoe JM; Pomerantz JL (2016). "NK Cell Maturation and Cytotoxicity Are Controlled by the Intramembrane Aspartyl Protease SPPL3". Journal of Immunology. 196 (6): 2614–26. doi:10.4049/jimmunol.1501970. PMID 26851218.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Voss M, Fukumori A, Kuhn PH, Künzel U, Klier B, Grammer G, Haug-Kröper M, Kremmer E, Lichtenthaler SF, Steiner H, Schröder B, Haass C, Fluhrer R (2012). "Foamy virus envelope protein is a substrate for signal peptide peptidase-like 3 (SPPL3)". The Journal of Biological Chemistry. 287 (52): 43401–9. doi:10.1074/jbc.M112.371369. PMC 3527927. PMID 23132852.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Mironov AA (2014). "ER-Golgi transport could occur in the absence of COPII vesicles". Nat Rev Mol Cell Biol. 15 (3): 1. doi:10.1038/nrm3588-c1. PMID 24496389.
- ^ a b Beznoussenko GV, Parashuraman S, Rizzo R, Polishchuk R, Martella O, Di Giandomenico D, Fusella A, Spaar A, Sallese M, Capestrano MG, Pavelka M, Vos MR, Rikers YG, Helms V, Mironov AA, Luini A (2014). "Transport of soluble proteins through the Golgi occurs by diffusion via continuities across cisternae". eLife. 3: e02009. doi:10.7554/eLife.02009. PMID 24867214.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ anon. "The Nobel Prize in Physiology or Medicine 2013". www.nobelprize.org. Alfred Nobel Memorial Foundation. Retrieved 7 April 2017.
- ^ Schekman, Randy. "Nobel Prize Lecture". YouTube. Nobel Prize YouTube Channel. Retrieved 8 April 2017.
- ^ Ge L, Zhang M, Schekman R (2014). "Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment". eLife. 3: e04135. doi:10.7554/eLife.04135. PMID 25432021.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Zanetti G, Prinz S, Daum S, Meister A, Schekman R, Bacia K, Briggs JA. (2013). "The structure of the COPII transport-vesicle coat assembled on membranes". eLife. 2: e00951. doi:10.7554/eLife.00951. PMID 24062940.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Santos AJ, Raote I, Scarpa M, Brouwers N, Malhotra V (2015). "TANGO1 recruits ERGIC membranes to the endoplasmic reticulum for procollagen export". eLife. 4: e10982. doi:10.7554/eLife.10982. PMID 26568311.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Zong M, Satoh A, Yu MK, Siu KY, Ng WY, Chan HC, Tanner JA, Yu S (2012). "TRAPPC9 mediates the interaction between p150 and COPII vesicles at the target membrane". PLoS One. 7: e29995. doi:10.1371/journal.pone.0029995. PMID 22279557.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ De Matteis MA, Rega LR (2015). "Endoplasmic reticulum-Golgi complex membrane contact sites". Curr Opin Cell Biol. 35: 43–50. doi:10.1016/j.ceb.2015.04.001. PMID 25950841.
- ^ Kurokawa,, K; Okamoto,, M; Nakano, A. "A model of ER-Golgi cargo transport by a 'hug-and-kiss' action of cis-Golgi". ncbi.nlm.nih.gov/. Nature Communications. Retrieved 3 June 2017.
{{cite web}}: CS1 maint: extra punctuation (link) - ^ Kurokawa K, Okamoto M, Nakano A (2014). "Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER". Nat Commun. 5: 3653. doi:10.1038/ncomms4653. PMID 24728174.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Burman JL, Hamlin JN, McPherson PS (2010). "Scyl1 regulates Golgi morphology". PLoS One. 5 (3): e9537. doi:10.1371/journal.pone.0009537. PMID 20209057.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Lavieu G, Zheng H, Rothman JE (2013). "Stapled Golgi cisternae remain in place as cargo passes through the stack". eLife. 2: e00558. doi:10.7554/eLife.00558. PMC 3673335. PMID 23755362.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Dancourt J, Zheng H, Bottanelli F, Allgeyer ES, Bewersdorf J, Graham M, Liu X, Rothman JE, Lavieu G (2016). "Small cargoes pass through synthetically glued Golgi stacks". FEBS Letters. 590 (12): 1675–86. doi:10.1002/1873-3468.12210. PMC 4925213. PMID 27174538.
- ^ Pellett PA, Dietrich F, Bewersdorf J, Rothman JE, Lavieu G. (2013). "Inter-Golgi transport mediated by COPI-containing vesicles carrying small cargoes". eLife. 2: e01296. doi:10.7554/eLife.01296. PMID 24137546.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Tie HC, Mahajan D, Chen B, Cheng L, VanDongen AM, Lu L (2016). "A novel imaging method for quantitative Golgi localization reveals differential intra-Golgi trafficking of secretory cargoes". Molecular Biology of the Cell. 27 (5): 848–61. doi:10.1091/mbc.E15-09-0664. PMC 4803310. PMID 26764092.
- ^ Sachdeva H, Barma M, Rao M (2016). "Nonequilibrium description of de novo biogenesis and transport through Golgi-like cisternae" (PDF). Scientific Reports. 6: 38840. doi:10.1038/srep38840. PMC 5171829. PMID 27991496.
- ^ Fraisier V, Kasri A, Miserey-Lenkei S, Sibarita JB, Nair D, Mayeux A, Bardin S, Toyoda Y, Poser I, Poznyakovskiy A, Goud B, Hyman AA, Dimitrov A (2013). "C11ORF24 is a novel type I membrane protein that cycles between the Golgi apparatus and the plasma membrane in Rab6-positive vesicles". Plos One. 8 (12): e82223. doi:10.1371/journal.pone.0082223. PMC 3846831. PMID 24312644.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Chiang L, Karvar S, Hamm-Alvarez SF (2012). "Direct imaging of RAB27B-enriched secretory vesicle biogenesis in lacrimal acinar cells reveals origins on a nascent vesicle budding site". Plos One. 7 (2): e31789. doi:10.1371/journal.pone.0031789. PMC 3282733. PMID 22363735.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Clermont Y, Rambourg A, Hermo L. (1995). "Trans-Golgi network (TGN) of different cell types: three-dimensional structural characteristics and variability". Anat Rec. 242 (3): 289–301. doi:10.1002/ar.1092420302. PMID 7573976.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Bonifacino JS (2014). "Adaptor proteins involved in polarized sorting". The Journal of Cell Biology. 204 (1): 7–17. doi:10.1083/jcb.201310021. PMC 3882786. PMID 24395635.
- ^ Cao X, Surma MA, Simons K (2012). "Polarized sorting and trafficking in epithelial cells". Cell Research. 22 (5): 793–805. doi:10.1038/cr.2012.64. PMC 3343658. PMID 22525333.
- ^ Tenorio MJ, Luchsinger C, Mardones GA, PLoS One. 2015 Aug 10;10(8):e0135260. doi: 10.1371/journal.pone.0135260. "Protein kinase A activity is necessary for fission and fusion of Golgi to endoplasmic reticulum retrograde tubules". journals.plos.org/. PLOS. Retrieved 5 May 2017.
{{cite web}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Taverna E, Mora-Bermúdez F, Strzyz PJ, Florio M, Icha J, Haffner C, Norden C, Wilsch-Bräuninger M, Huttner WB (2016). "Non-canonical features of the Golgi apparatus in bipolar epithelial neural stem cells". Scientific Reports. 6: 21206. doi:10.1038/srep21206. PMC 4754753. PMID 26879757.
- ^ Sengupta P, Satpute-Krishnan P, Seo AY, Burnette DT, Patterson GH, Lippincott-Schwartz J (2015). "ER trapping reveals Golgi enzymes continually revisit the ER through a recycling pathway that controls Golgi organization". Proceedings of the National Academy of Sciences of the United States of America. 112 (49): E6752–61. doi:10.1073/pnas.1520957112. PMC 4679030. PMID 26598700.
- ^ Guichet PO, Guelfi S, Ripoll C, Teigell M, Sabourin JC, Bauchet L, Rigau V, Rothhut B, Hugnot JP (2016). "Asymmetric Distribution of GFAP in Glioma Multipotent Cells". Plos One. 11 (3): e0151274. doi:10.1371/journal.pone.0151274. PMC 4783030. PMID 26953813.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Cervigni RI, Bonavita R, Barretta ML, Spano D, Ayala I, Nakamura N, Corda D, Colanzi A (2015). "JNK2 controls fragmentation of the Golgi complex and the G2/M transition through phosphorylation of GRASP65". Journal of Cell Science. 128 (12): 2249–60. doi:10.1242/jcs.164871. PMID 25948586.
- ^ Goberdhan DC, Wilson C, Harris AL (2016). "Amino Acid Sensing by mTORC1: Intracellular Transporters Mark the Spot". Cell Metabolism. 23 (4): 580–9. doi:10.1016/j.cmet.2016.03.013. PMC 5067300. PMID 27076075.
- ^ Cardenas J, Rivero S, Goud B, Bornens M, Rios RM (2009). "Golgi localisation of GMAP210 requires two distinct cis-membrane binding mechanisms". BMC Biology. 7: 56. doi:10.1186/1741-7007-7-56. PMC 2744908. PMID 19715559.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Biazik J, Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2015). "Ultrastructural relationship of the phagophore with surrounding organelles". Autophagy. 11 (3): 439–51. doi:10.1080/15548627.2015.1017178. PMC 4502653. PMID 25714487.
- ^ a b Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B (2013). "Organization and function of membrane contact sites". Biochimica Et Biophysica Acta. 1833 (11): 2526–41. doi:10.1016/j.bbamcr.2013.01.028. PMID 23380708.
- ^ a b Olkkonen VM (2015). "OSBP-Related Protein Family in Lipid Transport Over Membrane Contact Sites". Lipid Insights. 8 (Suppl 1): 1–9. doi:10.4137/LPI.S31726. PMC 4685180. PMID 26715851.
- ^ Yamaguchi H, Arakawa S, Kanaseki T, Miyatsuka T, Fujitani Y, Watada H, Tsujimoto Y, Shimizu S (2016). "Golgi membrane-associated degradation pathway in yeast and mammals". The EMBO Journal. 35 (18): 1991–2007. doi:10.15252/embj.201593191. PMC 5282831. PMID 27511903.
{{cite journal}}: Cite has empty unknown parameter:|1=(help); Unknown parameter|embargo=ignored (|pmc-embargo-date=suggested) (help) - ^ Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J (2017). "Applying systems-level spectral imaging and analysis to reveal the organelle interactome". Nature. doi:10.1038/nature22369. PMID 28538724.
- ^ a b c Spooner RA, Lord JM (2015). "Ricin trafficking in cells". Toxins. 7 (1): 49–65. doi:10.3390/toxins7010049. PMC 4303813. PMID 25584427.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Mesmin B, Antonny B (2016). "The counterflow transport of sterols and PI4P". Biochimica Et Biophysica Acta. 1861 (8 Pt B): 940–51. doi:10.1016/j.bbalip.2016.02.024. PMID 26928592.
- ^ a b Patterson GH, Hirschberg K, Polishchuk RS, Gerlich D, Phair RD, Lippincott-Schwartz J (2008). "Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system" (PDF). Cell. 133 (6): 1055–67. doi:10.1016/j.cell.2008.04.044. PMC 2481404. PMID 18555781.
- ^ a b Smits P, Bolton AD, Funari V, Hong M, Boyden ED, Lu L, Manning DK, Dwyer ND, Moran JL, Prysak M, Merriman B, Nelson SF, Bonafé L, Superti-Furga A, Ikegawa S, Krakow D, Cohn DH, Kirchhausen T, Warman ML, Beier DR (2010). "Lethal skeletal dysplasia in mice and humans lacking the golgin GMAP-210". The New England Journal of Medicine. 362 (3): 206–16. doi:10.1056/NEJMoa0900158. PMC 3108191. PMID 20089971.
- ^ Passemard S, Perez F, Colin-Lemesre E, Rasika S, Gressens P, El Ghouzzi V (2017). "Golgi trafficking defects in postnatal microcephaly: The evidence for "Golgipathies"". Progress in Neurobiology. 153: 46–63. doi:10.1016/j.pneurobio.2017.03.007. PMID 28377289.
- ^ Potelle S, Klein A, Foulquier F (2015). "Golgi post-translational modifications and associated diseases". Journal of Inherited Metabolic Disease. 38 (4): 741–51. doi:10.1007/s10545-015-9851-7. PMID 25967285.
- ^ Kim S, Hill A, Warman ML, Smits P (2012). "Golgi disruption and early embryonic lethality in mice lacking USO1". Plos One. 7 (11): e50530. doi:10.1371/journal.pone.0050530. PMC 3503957. PMID 23185636.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ (2008). "The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex". PLoS Genetics. 4 (12): e1000315. doi:10.1371/journal.pgen.1000315. PMC 2602600. PMID 19112494.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Litterman N, Ikeuchi Y, Gallardo G, O'Connell BC, Sowa ME, Gygi SP, Harper JW, Bonni A (2011). "An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning". Plos Biology. 9 (5): e1001060. doi:10.1371/journal.pbio.1001060. PMC 3091842. PMID 21572988.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Dbouk HA, Mroue RM, El-Sabban ME, Talhouk RS (2009). "Connexins: a myriad of functions extending beyond assembly of gap junction channels". Cell Communication and Signaling : CCS. 7: 4. doi:10.1186/1478-811X-7-4. PMC 2660342. PMID 19284610.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Xu H, Su W, Cai M, Jiang J, Zeng X, Wang H (2013). "The asymmetrical structure of Golgi apparatus membranes revealed by in situ atomic force microscope". Plos One. 8 (4): e61596. doi:10.1371/journal.pone.0061596. PMC 3628984. PMID 23613878.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Cheung PY, Limouse C, Mabuchi H, Pfeffer SR (2015). "Protein flexibility is required for vesicle tethering at the Golgi". eLife. 4. doi:10.7554/eLife.12790. PMC 4721967. PMID 26653856.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Tan X, Cao K, Liu F, Li Y, Li P, Gao C, Ding Y, Lan Z, Shi Z, Rui Q, Feng Y, Liu Y, Zhao Y, Wu C, Zhang Q, Li Y, Jiang L, Bao Y (2016). "Arabidopsis COG Complex Subunits COG3 and COG8 Modulate Golgi Morphology, Vesicle Trafficking Homeostasis and Are Essential for Pollen Tube Growth". PLoS Genetics. 12 (7): e1006140. doi:10.1371/journal.pgen.1006140. PMC 4957783. PMID 27448097.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Patwardhan A, Bardin S, Miserey-Lenkei S, Larue L, Goud B, Raposo G, Delevoye C (2017). "Routing of the RAB6 secretory pathway towards the lysosome related organelle of melanocytes". Nature Communications. 8: 15835. doi:10.1038/ncomms15835. PMID 28607494.
External links[edit]
- Three iBiology seminars by Jennifer Lippincott-Schwartz on YouTube:
- Three iBiology seminars by Kai Simons on YouTube:
- Three iBiology seminars by Ronald Vale on YouTube:









