SpoT
Bifunctional (p)ppGpp synthase/hydrolase SpoT or SpoT is a regulatory enzyme in the RelA/SpoT Homologue (RSH) protein family that synthesizes and hydrolyzes (p)ppGpp to regulate the bacterial stringent response to environmental stressors.[3] SpoT is considered a "long" form RSH protein and is found in many bacteria and plant chloroplasts.[4] SpoT and its homologues have been studied in bacterial model organism E.coli for their role in the production and degradation of (p)ppGpp in the stringent response pathway.
| Bifunctional (p)ppGpp synthase/hydrolase SpoT | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | spoT | ||||||
| UniProt | P0AG24 | ||||||
| |||||||
Role in Stringent Response Pathway[edit]
The stringent response regulated by SpoT, RelA, and their homologues can cause a bacterium to increase its persistence in stressful environments.[5] SpoT can act as both a hydrolase and a synthetase to (p)ppGpp alarmones in the stringent response pathway with Mn2+ as its cofactor.[6] When there are environmental stressors present, SpoT uses ATP and GDP to synthesize (p)ppGpp and catalyze the stringent response.[7] When stressors are removed and a stringent response is no longer necessary SpoT hydrolyzes (p)ppGpp, cleaving it into GTP and diphosphate.[7] Environmental stressors including but not limited to amino acid starvation,[3] carbon deficiencies,[8] phosphate deficiencies[9] and changes in temperature[4] have been documented to cause the gene encoding SpoT to activate.
The acyl carrier protein (ACP) binds to the TGS domain of SpoT; this binding is probably influenced by the ratio of unacylated ACP to acylated ACP in the cell.[10]
Role as a Hydrolase[edit]
SpoT mainly serves as a hydrolase in systems similar to E.coli. SpoT's hydrolase activity is Mn2+-dependent with a conserved His-Asp (HD) motif.[11] Phosphate starvation is sensed by SpoT hydrolase to elevate (p)ppGpp, which induces IraP, a RssB antiadaptor that antagonizes RssB activation of RpoS turnover, thereby inducing RpoS.[9]
SpoT in E. coli[edit]
In E. coli, the SpoT protein consists of 702 amino acids.[6] E.coli uses RelA and SpoT as its two main (p)ppGpp regulating enzymes.[12] When the gene for encoding RelA is nonfunctional, E. coli can still regulate (p)ppGpp through SpoT as it has both HD and SYNTH domains.
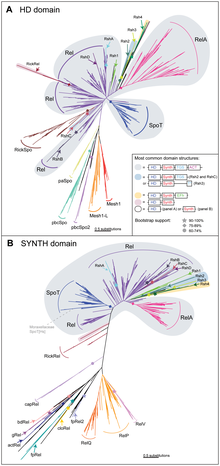
Related Proteins and the RelA/SpoT Homologue Superfamily[edit]
SpoT and RelA have many homologous variations, forming the RelA/SpoT Homologue (RSH) protein family. These homologues serve similar functions to SpoT and RelA in stringent responses. Protein domains observed in members of the RSH protein family are separated by hydrolase (HD) functionality and synthetase (SYNTH) functionality (see Figure 2).[4]
References[edit]
- ^ Maxwell A (2021-07-28). "Faculty Opinions recommendation of Highly accurate protein structure prediction with AlphaFold". doi:10.3410/f.740477161.793587439. S2CID 242530331.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. (January 2022). "AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models". Nucleic Acids Research. 50 (D1): D439–D444. doi:10.1093/nar/gkab1061. PMC 8728224. PMID 34791371.
- ^ a b Pausch P, Abdelshahid M, Steinchen W, Schäfer H, Gratani FL, Freibert SA, et al. (September 2020). "Structural Basis for Regulation of the Opposing (p)ppGpp Synthetase and Hydrolase within the Stringent Response Orchestrator Rel". Cell Reports. 32 (11): 108157. doi:10.1016/j.celrep.2020.108157. PMID 32937119. S2CID 221770084.
- ^ a b c d Atkinson GC, Tenson T, Hauryliuk V (2011-08-09). "The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life". PLOS ONE. 6 (8): e23479. Bibcode:2011PLoSO...623479A. doi:10.1371/journal.pone.0023479. PMC 3153485. PMID 21858139.
- ^ Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, et al. (July 2019). "Definitions and guidelines for research on antibiotic persistence". Nature Reviews. Microbiology. 17 (7): 441–448. doi:10.1038/s41579-019-0196-3. PMC 7136161. PMID 30980069.
- ^ a b "spoT - Bifunctional (p)ppGpp synthase/hydrolase SpoT - Escherichia coli (strain K12) - spoT gene & protein". www.uniprot.org. Retrieved 2022-05-01.
- ^ a b Ronneau S, Hallez R (July 2019). "Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria". FEMS Microbiology Reviews. 43 (4): 389–400. doi:10.1093/femsre/fuz009. PMC 6606846. PMID 30980074.
- ^ Lesley JA, Shapiro L (October 2008). "SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus". Journal of Bacteriology. 190 (20): 6867–6880. doi:10.1128/JB.00700-08. PMC 2566184. PMID 18723629.
- ^ a b Spira B, Silberstein N, Yagil E (July 1995). "Guanosine 3',5'-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi". Journal of Bacteriology. 177 (14): 4053–4058. doi:10.1128/jb.177.14.4053-4058.1995. PMC 177136. PMID 7608079.
- ^ Battesti A, Bouveret E (November 2006). "Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism". Molecular Microbiology. 62 (4): 1048–1063. doi:10.1111/j.1365-2958.2006.05442.x. PMID 17078815. S2CID 7857443.
- ^ Yakunin AF, Proudfoot M, Kuznetsova E, Savchenko A, Brown G, Arrowsmith CH, Edwards AM (August 2004). "The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2',3'-cyclic phosphodiesterase, 2'-nucleotidase, and phosphatase activities". The Journal of Biological Chemistry. 279 (35): 36819–36827. doi:10.1074/jbc.M405120200. PMID 15210699.
- ^ Liu K, Bittner AN, Wang JD (April 2015). "Diversity in (p)ppGpp metabolism and effectors". Current Opinion in Microbiology. Cell regulation. 24: 72–79. doi:10.1016/j.mib.2015.01.012. PMC 4380541. PMID 25636134.
Further reading[edit]
- Srivatsan A, Wang JD (April 2008). "Control of bacterial transcription, translation and replication by (p)ppGpp". Current Opinion in Microbiology. 11 (2): 100–5. doi:10.1016/j.mib.2008.02.001. PMID 18359660.
- Wendrich TM, Marahiel MA (October 1997). "Cloning and characterization of a relA/spoT homologue from Bacillus subtilis". Molecular Microbiology. 26 (1): 65–79. doi:10.1046/j.1365-2958.1997.5511919.x. PMID 9383190. S2CID 33335651.
