User:Kmcke14/sandbox2
New Sandbox for Mass Spec Class
 | This is a user sandbox of Kmcke14. You can use it for testing or practicing edits. This is not the sandbox where you should draft your assigned article for a dashboard.wikiedu.org course. To find the right sandbox for your assignment, visit your Dashboard course page and follow the Sandbox Draft link for your assigned article in the My Articles section. |


Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS).[1] Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them.[2] ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis.[3] Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact.[4] The technique only works well for higher charge state ions (z>2).[2] However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins.[5] This makes the technique important for top-down proteomics.The method was developed by Hunt and coworkers at the University of Virginia.[6]
History[edit]
Electron-capture dissociation (ECD) was developed in 1998 to fragment large proteins for mass spectrometric analysis.[7] Because ECD requires a large amount of near-thermal electrons (<0.2eV), originally it was used exclusively with Fourier transform ion cyclotron resonance mass spectrometry (FTICR), the most expensive form of MS instrumentation.[8] Less costly options such as quadrupole time-of-flight (Q-TOF), quadrupole ion trap (QIT) and linear quadrupole ion trap (QLT) instruments used the more energy-intensive collision-induced dissociation method (CID), resulting in random fragmentation of peptides and proteins.[9] In 2004 Syka et al announced the creation of ETD, a dissociation method similar to ECD, but using a low-cost, widely available commercial spectrometer. The first ETD experiments were run on a QLT mass spectrometer with an electrospray ionization (ESI) source.[10]
Principle of operation[edit]
Several steps are involved in electron transfer dissociation. Usually a protein mixture is first separated using high performance liquid chromatography (HPLC). Next multiply-protonated precursor molecules are generated by electrospray ionization and injected into the mass spectrometer. (Only molecules with a charge of 2+ or greater can be used in ETD.) In order for an electron to be transferred to the positive precursor molecules radical anions are generated and put into the ion trap with them. During the ion/ion reaction an electron is transferred to the positively-charged protein or peptide, causing fragmentation along the peptide backbone. Finally the resultant fragments are mass analyzed.[11]
Radical anion preparation[edit]
In the original ETD experiments anthracene (C14H10) was was used to generate reactive radical anions through negative chemical ionization.[10] Several polycyclic aromatic hydrocarbon molecules have been used in subsequent experiments, with fluoranthene currently the preferred reagent.[12] Fluoranthene has only about 40% efficiency in electron transfer, however, so other molecules with low electron affinity are being sought.[11]
Injection and fragmentation[edit]

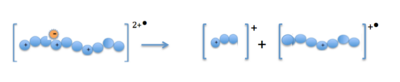
When the precursor cations (proteins or peptides) and radical anions are combined in the ion trap an electron is transferred to the mulitply-charged cation. This forms an unstable positive radical cation with one less positive charge and an odd electron.[13] Fragmentation takes place along the peptide backbone at a N− Cα bond, resulting in c- and z-type fragment ions.[3]
Mass analysis[edit]
Fragmentation caused by ETD allows more complete protein sequence information to be obtained from ETD spectra than from CID tandem mass spectrometry. Because many peptide backbone c- and z- type ions are detected, almost complete sequence coverage of many peptides can be discerned from ETD fragmentation spectra.[14] Sequences of 15-40 amino acids at both the N-terminus and the C-terminus of the protein can be read using mass-to-charge values for the singly and doubly charged ions. These sequences, together with the measured mass of the intact protein, can be compared to database entries for known proteins and to reveal post-translational modifications.[15]
Instrumentation[edit]

Electron transfer dissociation takes place in an ion trap mass spectrometer with an electrospray ionization source. The first ETD experiments at the University of Virginia utilized a radio frequency quadrupole linear ion trap (LQT) modified with a chemical ionization (CI) source at the back side of the instrument (see diagram at right).[10] Because a spectrum can be obtained in about 300 milliseconds, liquid chromatography is often coupled with the ETD MS/MS.[11] The disadvantage of using LQT is that the mass resolving power is less that that of other mass spectrometers.[5]
Subsequent studies have tried different instrumentation to improve mass resolution. In 2006 a group at Perdue University used a quadrupole/time-of-flight (QqTOF) tandem mass spectrometer with pulsed nano-ESI/atmospheric pressure chemical ionization (APCI) dual ionization source using radical anions of 1,3-dinitrobenzene as the electron donor.[16]
Proteomics[edit]
ETD is advantageous for the fragmentation of longer peptides or even entire proteins.
For biopharmaceutical characterization, ETD is a powerful fragmentation technique for determining modification sites of labile post-translational modifications (PTMs), which are often difficult to characterize using CID. (also waters page)
Changes to the method[edit]
newer applications including characterization of PTMs, non-tryptic peptides and intact proteins. (Kim Review) lectron-transfer and higher-energy collision dissociation (EThcD) is a combination ETD and HCD where the peptide precursor is initially subjected to an ion/ion reaction with fluoranthene anions in a linear ion trap, which generates c- and z-ions.[17] In the second step HCD all-ion fragmentation is applied to all ETD derived ions to generate b- and y- ions prior to final analysis in the orbitrap analyzer.[18] This method employs dual fragmentation to generate ion- and thus data-rich MS/MS spectra for peptide sequencing and PTM localization.[19]
See also[edit]
References[edit]
- ^ Dass, Chhabil (2007). Fundamentals of Contemporary Mass Spectrometry. Hoboken, New Jersey: John Wiley & Sons. p. 128. ISBN 978-0-470-11848-1.
- ^ a b Hart-Smith, Gene (2014-01-15). "A review of electron-capture and electron-transfer dissociation tandem mass spectrometry in polymer chemistry". Analytica Chimica Acta. Polymer Mass Spectrometry. 808: 44–55. doi:10.1016/j.aca.2013.09.033.
- ^ a b Brodbelt, Jennifer S. (2015-12-11). "Ion Activation Methods for Peptides and Proteins". Analytical Chemistry. 88 (1): 30–51. doi:10.1021/acs.analchem.5b04563.
- ^ Coon, Joshua J.,; Syka, John E.P.; Shabanowitz, Jeffrey; Hunt, Donald F. (April 2005). "Tandem Mass Spectrometry for Peptide and Protein Sequence Analysis". BioTechniques. PMID 15884666. Retrieved April 15, 2016.
{{cite journal}}: CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link) - ^ a b Qi, Yulin; Volmer, Dietrich A. (2015-10-01). "Electron-based fragmentation methods in mass spectrometry: An overview". Mass Spectrometry Reviews: n/a–n/a. doi:10.1002/mas.21482. ISSN 1098-2787.
- ^ US patent 7534622, Donald F. Hunt, Joshua J. Coon, John E.P. Syka, Jarrod A. Marto, "Electron transfer dissociation for biopolymer sequence mass spectrometric analysis", issued 2009-05-19
- ^ Zubarev, Roman A.; Kelleher, Neil L.; McLafferty, Fred W. (1998-04-01). "Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process". Journal of the American Chemical Society. 120 (13): 3265–3266. doi:10.1021/ja973478k. ISSN 0002-7863.
- ^ McLafferty, Fred W.; Horn, David M.; Breuker, Kathrin; Ge, Ying; Lewis, Mark A.; Cerda, Blas; Zubarev, Roman A.; Carpenter, Barry K. (2001-03-01). "Electron capture dissociation of gaseous multiply charged ions by Fourier-transform ion cyclotron resonance". Journal of the American Society for Mass Spectrometry. 12 (3): 245–249. doi:10.1016/S1044-0305(00)00223-3. ISSN 1044-0305.
- ^ Mitchell Wells, J.; McLuckey, Scott A. (2005-01-01). Enzymology, BT - Methods in (ed.). Collision‐Induced Dissociation (CID) of Peptides and Proteins. Biological Mass Spectrometry. Vol. 402. Academic Press. pp. 148–185. doi:10.1016/s0076-6879(05)02005-7.
- ^ a b c Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF (2004). "Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry". Proc. Natl. Acad. Sci. U.S.A. 101 (26): 9528–33. Bibcode:2004PNAS..101.9528S. doi:10.1073/pnas.0402700101. PMC 470779. PMID 15210983.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Kim, Min-Sik; Pandey, Akhilesh (2012-02-01). "Electron transfer dissociation mass spectrometry in proteomics". PROTEOMICS. 12 (4–5): 530–542. doi:10.1002/pmic.201100517. ISSN 1615-9861. PMC 3664229. PMID 22246976.
- ^ Chi, An; Huttenhower, Curtis; Geer, Lewis Y.; Coon, Joshua J.; Syka, John E. P.; Bai, Dina L.; Shabanowitz, Jeffrey; Burke, Daniel J.; Troyanskaya, Olga G. (2007-02-13). "Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry". Proceedings of the National Academy of Sciences. 104 (7): 2193–2198. doi:10.1073/pnas.0607084104. ISSN 0027-8424. PMC 1892997. PMID 17287358.
- ^ "Electron Transfer Dissociation". The National High Magnetic Field Laboratory. August 28, 2015. Retrieved March 1, 2016.
- ^ Zhang, Qibin; Frolov, Andrej; Tang, Ning; Hoffmann, Ralf; van de Goor, Tom; Metz, Thomas O.; Smith, Richard D. (2007-03-15). "Application of electron transfer dissociation mass spectrometry in analyses of non-enzymatically glycated peptides". Rapid Communications in Mass Spectrometry. 21 (5): 661–666. doi:10.1002/rcm.2884. ISSN 1097-0231. PMC 2731431. PMID 17279487.
- ^ Chi, An; Bai, Dina L.; Geer, Lewis Y.; Shabanowitz, Jeffrey; Hunt, Donald F. (2007-01-01). "Analysis of intact proteins on a chromatographic time scale by electron transfer dissociation tandem mass spectrometry". International Journal of Mass Spectrometry. Donald F. Hunt Honour Issue. 259 (1–3): 197–203. doi:10.1016/j.ijms.2006.09.030. PMC 1826913. PMID 17364019.
- ^ Xia, Yu; Chrisman, Paul A.; Erickson, David E.; Liu, Jian; Liang, Xiaorong; Londry, Frank A.; Yang, Min J.; McLuckey, Scott A. (2006-06-01). "Implementation of Ion/Ion Reactions in a Quadrupole/Time-of-Flight Tandem Mass Spectrometer". Analytical Chemistry. 78 (12): 4146–4154. doi:10.1021/ac0606296. ISSN 0003-2700. PMC 2575740. PMID 16771545.
- ^ Frese, Christian K.; A. F. Maarten Altelaar; Henk van den Toorn; Dirk Nolting; Jens Griep-Raming; Albert J. R. Heck; Shabaz Mohammed (November 20, 2012). "Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry". Anal. Chem. 84 (22): 9668–73. doi:10.1021/ac3025366. PMID 23106539.
- ^ Olsen JV; Macek B; Lange O; Makarov A; Horning S; Mann M (September 2007). "Higher-energy C-trap dissociation for peptide modification analysis". Nature Methods. 4 (9): 709–712. doi:10.1038/nmeth1060. PMID 17721543.
- ^ Frese, Christian K.; Houjiang Zhou; Thomas Taus; A. F. Maarten Altelaar; Karl Mechtler; Albert J. R. Heck; Shabaz Mohammed (March 1, 2013). "Unambiguous Phosphosite Localization using Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)". J Proteome Res. 12 (3): 1520–5. doi:10.1021/pr301130k. PMC 3588588. PMID 23347405.

