Talk:Pseudogene
 | Ideal sources for Wikipedia's health content are defined in the guideline Wikipedia:Identifying reliable sources (medicine) and are typically review articles. Here are links to possibly useful sources of information about Pseudogene.
|
| It is requested that a biology diagram or diagrams be included in this article to improve its quality. Specific illustrations, plots or diagrams can be requested at the Graphic Lab. For more information, refer to discussion on this page and/or the listing at Wikipedia:Requested images. |
| This article is rated B-class on Wikipedia's content assessment scale. It is of interest to the following WikiProjects: | |||||||||||||||||
| |||||||||||||||||
RNAi[edit]
I don't think this sentence is really accurate:
- The growing understanding of RNA interference likely means that some former pseudogenes may be reclassified as RNAi sequences.
To my understanding, RNAi coming from the genome means miRNA, and you need very specific stem-loop structures in miRNA genes that won't arise from a pseudogene any more often than from anywhere else. The Hirotsune paper discusses pseudo-mRNA mediation of a real mRNA, but not through the same degradation machinery that (I believe) "RNAi" refers to.
--Mike Lin 18:32, 18 January 2006 (UTC)
RNAi is not limited to miRNA. I have reworded that section and definition.Ted 16:25, 19 January 2006 (UTC)
- Can you please clarify for my own knowledge, what does "RNAi" encompass besides miRNA (naturally occurring in the genome) and siRNA (artificially introduced)? --Mike Lin 06:22, 20 January 2006 (UTC)
- My understanding of RNAi is any post transcriptional gene silencing (PTGS) involving dsRNA. It was first discovered adding anti-sense RNA which hybridized with the sense message and silenced expression. While the common use is siRNA with experimenter-derived dsRNA, it can also refer to anti-sense ssRNA binding with the sense strand forming dsRNA, and specialized sequences apparently designed for silencing (miRNA).Ted 23:49, 23 January 2006 (UTC)
It seems we are working off different definitions.
- Definition 1: A pseudogene does not produce a functional protein.
- Definition 2: A pseudogene does not produce a functional final product.
The problem with first definition is that some normal genes produce RNA as a final product, such as tRNA genes, etc. In addition, there are sequences that have homology with tRNA genes, but are not expressed. It takes some gymnastics to fit the pseudogene definition. Hirotsune (and the separate comments by Lee in the same issue of Nature) use this definition; further supported by Gerstein's lab (http://www.pseudogene.org). Many researchers use the generic "is never expressed."
The second definition is more expansive. Do a medline search for 'trna pseudogene' or 'rrna pseudogene' and you will find many researchers who use this term when the functional final product is an RNA. The mechanism for creating these sequences are the same as for the protein-coding pseudogenes (mainly duplicated), so it is hard to discount them entirely. Ted 18:05, 19 January 2006 (UTC)
- I agree with what you're saying, but do you have a suggestion for what to do? As you note, the definition of the term is ambiguous "in real life" among experts. I don't know if we can solve this on Wikipedia. --Mike Lin 06:26, 20 January 2006 (UTC)
- How about this?
- A pseudogene is a nucleotide sequences that is similar to a normal gene, but does not produce a functional final product. There are two variants. The first requires the final product to be a protein. The second allows the final product to be an RNA, such as tRNA or rRNA.Ted 14:08, 26 January 2006 (UTC)
Illegitimate addition to "Properties of Pseudogenes?[edit]
That section begins with the following: "Pseudogenes have two main functions; longevity of the genome life and enhancement of the gametes." I may be wrong, but something smells fishy there: perhaps a Creationist edit??? —Preceding unsigned comment added by 24.94.233.30 (talk) 06:08, 25 July 2009 (UTC)
- It sounds like a spoof. In any case, it is not sourced and it doesn't really fit in with what follows, so I will delete it. Boghog2 (talk) 14:27, 25 July 2009 (UTC)
Language of RNA Decoded: Study Reveals New Function for Pseudogenes and Noncoding RNAs[edit]
http://www.sciencedaily.com/releases/2010/06/100623132102.htm
Language of RNA Decoded: Study Reveals New Function for Pseudogenes and Noncoding RNAs
Now, a cancer genetics team at Beth Israel Deaconess Medical Center (BIDMC) suggests there is much more to RNA than meets the eye.
In a study appearing in the June 24, 2010 issue of Nature, the authors describe a new regulatory role for RNA -- independent of their protein-coding function -- that relies on their ability to communicate with one another. Of potentially even greater significance, because this new function also holds true for thousands of noncoding RNAs, the discovery dramatically increases the known pool of functional genetic information. 12.107.178.10 (talk) 19:01, 24 June 2010 (UTC) nobody
Please update this article.[edit]
Can someone please update this article. Some of these citations are over thirty years old. Listing Pseudogene as dysfunctional is just not the case anymore. — Preceding unsigned comment added by 108.65.170.183 (talk) 12:32, 15 April 2012 (UTC)
In support of this request:
Protein Coding 'Junk Genes' May Be Linked to Cancer
In the current paper in Nature Methods, researchers present a new proteogenomics method, which makes it possible to track down protein coding genes in the remaining 98.5% of the genome, something that until now has been an impossible task to pursue. Among other things, the research shows that some pseudogenes produce proteins indicating that they indeed have a function.
http://www.sciencedaily.com/releases/2013/11/131117155500.htm
-- Jo3sampl (talk) 13:57, 19 November 2013 (UTC)
I completely agree, "functionless" is just the wrong word to start with, for example.
Section about "Potential function" is confusing[edit]
This sections is confusing, because it a) refers to the definition/classification of pseudogenes, which does not fit there and b) the sentence says "This has led to the incorrect identification of pseudogenes. Examples include" before the list of potential functions. Probably the whole section before the list should be rewritten? In my opinion, a nice, recent overview of potential function is given in Pei 2012 [1] IronicPseudonyme (talk) 12:51, 27 January 2014 (UTC)
References
- ^ Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ, Harte R, Balasubramanian S, Tanzer A, Diekhans M, Reymond A, Hubbard TJ, Harrow J, Gerstein MB (2012). "The GENCODE pseudogene resource". Genome Biology. 13 (9): R51. doi:10.1186/gb-2012-13-9-r51. PMC 3491395. PMID 22951037.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
The image[edit]
I'm a new user. A question I asked at the Teahouse suggested that I come here to voice my concerns, so I hope I do it right. IMHO, the image on this article is pathetic. I'm an expert in the field, and I can only imagine that a novice looking at the imaqe would think "WTF?". Rather than satisfying the idea that "a picture is worth a thousand words", it would take me thousands of words to explain the "pathologies" in that image. The image was composed by a user that is no longer active. One of the Teahouse comments suggeted that I be bold and eliminate it. If I don't get objections here, that is what I'll do....and yes, I do plan to make new images and post them. DennisDrdfp (talk) 21:24, 28 December 2016 (UTC)
Thanks boghog. I just added my first image to the "processed" section. I intend to make more before deleting the original one.Drdfp (talk) 23:47, 28 December 2016 (UTC)
- There are a number of problems with this new figure. First of all, all the sources displayed are primary:
- Zhang X, Zhang J, Ping X, Wang QL, Lu X (2016). "Pseudogene transcripts: Participants in tumorigenicity and promising therapeutic targets". primary source. Leukemia Research. 42: 105–6. doi:10.1016/j.leukres.2015.12.011. PMID 26818436.
- Ganapathi MK, Jones WD, Sehouli J, Michener CM, Braicu IE, Norris EJ, Biscotti CV, Vaziri SA, Ganapathi RN (2016). "Expression profile of COL2A1 and the pseudogene SLC6A10P predicts tumor recurrence in high-grade serous ovarian cancer". primary source. International Journal of Cancer. 138 (3): 679–88. doi:10.1002/ijc.29815. PMID 26311224.
- Ye X, Fan F, Bhattacharya R, Bellister S, Boulbes DR, Wang R, Xia L, Ivan C, Zheng X, Calin GA, Wang J, Lu X, Ellis LM (2015). "VEGFR-1 Pseudogene Expression and Regulatory Function in Human Colorectal Cancer Cells". primary source. Molecular Cancer Research : MCR. 13 (9): 1274–82. doi:10.1158/1541-7786.MCR-15-0061. PMC 4573265. PMID 26041938.
- Zheng L, Li X, Gu Y, Lv X, Xi T (2015). "The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1". primary source. Breast Cancer Research and Treatment. 150 (1): 105–18. doi:10.1007/s10549-015-3298-2. PMID 25701119.
- Wikipedia prefers WP:SECONDARY sources, especially for medical related content (see WP:MEDRS). This is in part because a astonishingly high percentage of biomedical research cannot be repeated.
- Wikipedia should simply state facts. From WP:MEDMOS:
Do not hype a study by listing the names, credentials, institutions, or other "qualifications" of their authors. The text of the article should not needlessly duplicate the names, dates, titles, and other information about the source that you list in the citation.
- Per WP:NOTJOURNAL, wikipedia should not read like a scientific journal or research talk.
- Per WP:RECENT, the figure places too much emphasis on recent research. Again, it is much better to rely on secondary sources that review and summarize primary research. Boghog (talk) 08:52, 29 December 2016 (UTC)
- This may be a subtle but nevertheless an important point: Wikipedia articles should concentrate on the subject, not research on the subject. Boghog (talk) 13:32, 29 December 2016 (UTC)
A suggestion for the type of graphic that may be appropriate for the lead section is Pitman SD (November 2008). "Origin of pseudogenes". Pseudogenes. Not sure that I agree with the premise of this web site, but at least this figure looks uncontroversial. We cannot copy this figure without permission, but we could use it as a template for a new figure. Boghog (talk) 13:32, 29 December 2016 (UTC) Here is another figure that is very similar to the one above. Boghog (talk) 13:39, 29 December 2016 (UTC)
- @Boghog:I've looked at these images and others from the review articles you suggested elsewhere. I have reservations about all of them, and have given up on the possibility of having an informative, eye-catching image in the lead of the article, and will turn my attention to individual sections. DennisPietras (talk) 01:25, 4 January 2017 (UTC)
The pseudo-pseudogene section[edit]
I added it 12/28/2016Drdfp (talk) 22:55, 28 December 2016 (UTC)
Former first image deleted and revisions begun[edit]
Folks, I have completed a new first paragraph in what I hope to be an extensive update to this page, as suggested below. I appreciate any comments/suggestions, especially if you know how to insert explanatory footnotes "automatically". I asked for help with that in the teahouse, and haven't yet gotten a reply. I couldn't stand to look at that initial image anymore, so I deleted that. Thanks, Dennis Drdfp (talk) 04:15, 29 December 2016 (UTC)
- Hi Dennis, you might find the pages Help:Footnotes and Template:Ref helpful. (If you need further help or clarification, I'll be happy to help.) I have just a few minor suggestions: instead of using <i></i>, it's easier to use the wikimarkup (pairs of straight quotes: '' '') for italics. Footnotes should be in "wikivoice" (i.e. no signatures needed, no first/second person, etc.). Also, per WP:BOTTOMPOST, new talk page threads should go at the bottom of the page. Thank you for your contributions! Me, Myself & I (☮) (talk) 05:57, 29 December 2016 (UTC)
- I am not exactly clear what you mean by automatic insertion of explanatory footnotes. Underneath the edit window, if one changes <insert> to <Wiki markup>, a number of hyperlinked options appear including <ref></ref>. Press that, and the <ref></ref> characters will appear in the Wikitext. That will make it somewhat easier to insert footnotes. For inserting citations, one option is User:Diberri's Wikipedia template filling tool (instructions). Given a PubMed ID, one can quickly produce a full citation that can be copied and pasted into a Wikipedia article. In addition, this tool will add PMID, DOI, and PMC parameters that assist users in quickly finding the full article. Boghog (talk) 09:14, 29 December 2016 (UTC)
Please comment on this image[edit]
Please take a look at this image, which I extensively revised from a graphical abstract of a research paper. I believe that my edits make it "fair use" for inclusion in this article, in a section not yet written. I am interested in 1. whether you can understand it, 2. whether you think that it would be understandable by a typical reader, and 3. whether you think it should be in the article. Thanks DennisPietras (talk) 01:47, 4 January 2017 (UTC)
- The figure itself looks fine. However it should be accompanied by some text in body of the article (supported by an appropriate source of course) that refers to it. Also it probably should not be in the lead but in the cancer section. Finally I have increased the size slight so that it is legible without having to open it in a new window. Boghog (talk) 20:26, 4 January 2017 (UTC)
References
- boghogThanks for your comments, but I've had to remove it because it was deleted for not following wp guidelines for fair use or whatever. sigh. DennisPietras (talk) 04:50, 6 January 2017 (UTC)
Changes to last paragraph of pseudo-pseudogenes section[edit]
If I understand who edits things, I think that @Boghog: made changes to my original last paragraph. It is slowly sinking into my newbie brain that wp doesn't like direct quotes from publications even when they are cited. I thought that the changes made were a bit "cumbersome" or "difficult to follow", so I revised the last paragraph to, I think, improve it without using quotattions. DennisPietras (talk) 02:52, 4 January 2017 (UTC)
- No problem. I agree that your follow-up edits are an improvement. Cheers. Boghog (talk) 20:19, 4 January 2017 (UTC)
changes to unitary pseudogene section[edit]
Folks, I added a figure to this section and made several changes to the text to make it more accurate, IMHO. I noticed that the following 2 paragraphs:
- "Pseudogenes can complicate molecular genetic studies. For example, amplification of a gene by PCR may simultaneously amplify a pseudogene that shares similar sequences. This is known as PCR bias or amplification bias. Similarly, pseudogenes are sometimes annotated as genes in genome sequences.
- Processed pseudogenes often pose a problem for gene prediction programs, often being misidentified as real genes or exons. It has been proposed that identification of processed pseudogenes can help improve the accuracy of gene prediction methods.
were about properties of pseudogenes in general, not unitaries in particular, so I deleted them from this section and inserted them into the properties section. DennisPietras (talk) 17:09, 9 January 2017 (UTC)
end of unitary section - is this even correct???[edit]
Folks, The current last paragraph of the unitary section is "It has also been shown that the parent sequences that give rise to processed pseudogenes lose their coding potential faster than those giving rise to non-processed pseudogenes". I have read the reference cited fairly closely, and I think the last pararaph was inspired by this section of the paper:
- "In addition, 83.2% of processed and 79% of non-processed pseudogenes display disablements (defined as non-sense or frameshift mutations) in their putative ORFs, with average disablements of 6.2 per processed pseudogene and 2.4 per nonprocessed pseudogene. ...The differences in sequence identity and disablements between processed and nonprocessed pseudogenes are significant (P< 0.001, Wilcoxon rank-sum test), appearing to suggest that the sequences giving rise to processed pseudogenes lose coding potential more quickly than those for nonprocessed pseudogenes."
I believe that the last paragraph is misleading at best. Specifically "...the parent sequences that give rise to processed pseudogenes" could be replaced, logically, by "...the normal genes that give rise to processed pseudogenes". Doing that replacement would change the entire last paragraph to:
- "It has also been shown that the normal genes that give rise to processed pseudogenes lose their coding potential faster than those giving rise to non-processed pseudogenes".
There is no evidence of that as far as I know, and it certainly doesn't exist in the citation. I propose that the last line be changed to:
- "It has been shown that processed pseudogenes accumulate mutations faster than non-processed pseudogenes".
Comments? DennisPietras (talk) 17:53, 9 January 2017 (UTC)
- Since nobody objected, I made the change I suggested. DennisPietras (talk) 17:23, 12 January 2017 (UTC)
high mobility group discussion in the involvement in cancer section[edit]
Folks, whoever inserted the discussion cites "DeMartino et al 2016" repeatedly, yet the reference does not appear in the references section. I have tried to find it on medline, and have failed. I tried to use wikiblame (for only the second time) to find out who edited it, and it appears that the stuff was added by Glee1204, but there isn't even a user page by that name!!! What to do? Thanks, DennisPietras (talk) 01:24, 10 January 2017 (UTC)
- I just found the reference. It should be Esposito et al, 2014, yada, yada. I don't think there should be so much discussion of it, and I'm still concerned about why Glee1204 doesn't have a user page, so I'd still appreciate your comments. DennisPietras (talk) 01:51, 10 January 2017 (UTC)
- Userpages are optional; some editors go without one for their entire "career". It appears that Glee1204 only made one edit to mainspace, too. You could try to leave them a message on their talk page to see if they have anything to say about it, although they might not respond. If no one else objects, you can go ahead and boldly trim the section down. Me, Myself & I (☮) (talk)
- @Me, Myself, and I are Here:Thanks! Why Glee1204 decided to edit the first page I became interested in is a mystery. I've been thinking about adopting the bold approach. Thanks for the encouragement. DennisPietras (talk) 03:24, 11 January 2017 (UTC)
- Userpages are optional; some editors go without one for their entire "career". It appears that Glee1204 only made one edit to mainspace, too. You could try to leave them a message on their talk page to see if they have anything to say about it, although they might not respond. If no one else objects, you can go ahead and boldly trim the section down. Me, Myself & I (☮) (talk)
major revisions begun 1/12/2017[edit]
Folks, I've been thinking a lot about how to approach revisions in the rest of the article. Folks at the teahouse told me about the "in use" and "under construction" templates, and I intend to use them to let readers know that parts may be even messier for a while, but I'm not going to let this sit for long. I don't type fast. Fine motor control problems from MS, and yes, I realize that the world can see that and I don't care. sigh. So, please be patient. Thanks, DennisPietras (talk) 17:31, 12 January 2017 (UTC)
removed MKRN setion[edit]
I couldn't find anybody disputing the paper that dissed the mkrn-p1 report, so I deleted that section. DennisPietras (talk) 19:07, 12 January 2017 (UTC)
Removed Glee1204's insertion of "involvement in cancer"[edit]
Folks, in all of the long discussion of HMGA proteins, etc, there is one especially relevant statement: "Because of these pseudogenes' mutations, they can code for competitor proteins for the wild-type HMGA1 gene. Modification of HGMA1 expression results in modified chromatin remodeling and protein-protein (DeMartino et al., 2016)" I have spent more than enough of my life looking for that reference. It doesn't appear to exist. If I understand how to search who contributed what, Glee1204 added that entire section without adequate references, and that is the only edit Glee1204 has made on wiki. No user page for her/him. I have deleted that entire section. DennisPietras (talk) 03:05, 13 January 2017 (UTC)
sureys section deleted[edit]
Folks, one reference in that section was 10 years old and the other was mentioned earlier in the article. Plus, surveys doesn't fit under "funcions", where somebody placed it, so I deleted it entirely. DennisPietras (talk) 04:10, 13 January 2017 (UTC)
seminal RNAse discussion deleted[edit]
In brief, the discussion about seminal RNase is fubar and I have deleted it. If you want the gory details, read on.
The first citation to a paper about seminal RNase is this under gene ressurection: "The repair of lesions could be achieved by the reinsertion of a deleted segment, the removal (in frame) of an inserted segment, or other events that are likely to be improbable like gene conversion.[46]" 46 at that time was a reference from 2007 as follows:
- Sassi SO, Braun EL, Benner SA (April 2007). "The evolution of seminal ribonuclease: pseudogene reactivation or multiple gene inactivation events?". Molecular Biology and Evolution. 24 (4): 1012–24. doi:10.1093/molbev/msm020. PMID 17267422.
The next mention followed immediately after the statement above under the heading examples: "The bovine seminal ribonuclease, which had lain dormant for about 20 million years as a pseudogene, appears to have been resurrected by gene conversion.[47]" 47 at that time was a reference from 1996 as follows:
- Trabesinger-Ruef N, Jermann T, Zankel T, Durrant B, Frank G, Benner SA (March 1996). "Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function?". FEBS Letters. 382 (3): 319–22. doi:10.1016/0014-5793(96)00191-3. PMID 8605993.
Here's the almost unbelievable part: the assertion that the rnase gene was ressurected by gene conversion came from 1996, while the 2007 paper, which was mentioned first in "pseudogenes", actually convincingly (to me and the reviewers of a respected journal) showed that the rnase did not arise by gene conversion!!!!!
Can you say "fake news" DennisPietras (talk) 15:10, 13 January 2017 (UTC)
added gene conversion and deleted stuff about gene resurrection and examples[edit]
I added a discussion of pseudogenes and gene conversion in the "examples of pseudogene function" sections, which would "substitute" for a mention of gene resurection. The 4 references that remained in the "gene resurrection" and "examples" sections were from the last millenium, and not worth discussing, IMHO, so I deleted that discussion. DennisPietras (talk) 16:00, 13 January 2017 (UTC)
what was left of "transcription" became "bacterial pseudogenes"[edit]
almost done with the first pass!!!!!!!!!!!!!!!! DennisPietras (talk) 16:30, 13 January 2017 (UTC)
a knee slapping belly laugher! The "See also" section....[edit]
had a link
which lists 8 genes! 8-) DennisPietras (talk) 18:03, 13 January 2017 (UTC)
and so, it is finished....[edit]
no, of course it never will be, but I'm through with the construction signs! I raised it to B class and, because of all the indications that pseudogenes are involved in widespread gene regulation, raised it to high importance. This was more difficult and frustrating than I anticipated. Hopefully, Glee1204 won't buzz by and mess with it much. 8-) DennisPietras (talk) 20:45, 13 January 2017 (UTC)
Moved from article[edit]
Comments belong on the talk page, not in the article. Also it is rarely appropriate to mention dates, expect in a history section. Hence, I am moving the material here. Boghog (talk) 21:06, 13 January 2017 (UTC)
This section is added for the use of those who want to keep this article current, without messing up the organizational outline above, hint, hint..... Please help fight entropy!
- The role of PTENP1 in gastric cancer was investigated[1]
- A review of ceRNAs under the synonym "micro RNA sponges" appeared[2]
- It was found that low expression of RNA from a particular pseudogene was associated with longer time-to-recurrence after chemotherapy for some ovarian cancer.[3]
- It was reported that unusual translation initiation sites were used at the start of tumorigenesis.[4] This casts doubt on the idea that pseudogenes lacking the normal initiation site cannot be translated.
References
- ^ Guo X, Deng L, Deng K, Wang H, Shan T, Zhou H, Liang Z, Xia J, Li C (2016). "Pseudogene PTENP1 Suppresses Gastric Cancer Progression by Modulating PTEN". Anti-Cancer Agents in Medicinal Chemistry. 16 (4): 456–64. PMID 25968876.
- ^ Thomson DW, Dinger ME (May 2016). "Endogenous microRNA sponges: evidence and controversy". Nature Reviews. Genetics. 17 (5): 272–83. doi:10.1038/nrg.2016.20. PMID 27040487.
- ^ Ganapathi MK, Jones WD, Sehouli J, Michener CM, Braicu IE, Norris EJ, Biscotti CV, Vaziri SA, Ganapathi RN (February 2016). "Expression profile of COL2A1 and the pseudogene SLC6A10P predicts tumor recurrence in high-grade serous ovarian cancer". International Journal of Cancer. 138 (3): 679–88. doi:10.1002/ijc.29815. PMID 26311224.
- ^ Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, Weissman JS, Fuchs E (January 2017). "Translation from unconventional 5' start sites drives tumour initiation". Nature. doi:10.1038/nature21036. PMID 28077873.
lead figure[edit]
@Boghog: and everybody else: sure, it's strange, but so am I. Feel free to delete it if you think it's inappropriate, but I needed some silliness in my life. DennisPietras (talk) 21:35, 13 January 2017 (UTC)
- @Rahva.vaenlane: I reverted your deletion of the lead figure. As you can see, it passed review by several people who were very interested in the material on this page. I disagree with your interpretation. The figure shows damage to a gene by an ROS. Do you understand what that means? If you or anybody else doesn't, you or they can click on the ROS link in the caption to find out. The "bandage" on the gene evokes one of the most important points about pseudogenes. They are a bit injured, but they can still do stuff, and may be able to return to full function in time, just as people with bandages can and do. Also, the green beard, as I've linked to in the caption, is a full-fledged concept of it's own, which you or anybody else who doesn't know that can learn by following the link. Finally, I'm a newbie and may be wrong, but aren't you supposed to check the talk page before making a substantial deletion, to see what the consensus is? DennisPietras (talk) 19:29, 4 February 2017 (UTC)
- @DennisPietras and Rahva.vaenlane: Ideally the graphic in the lead should provide an overview of the subject. I suggested above that we use a figure from a review article. Since it copyrighted, we can not use it directly, but I have created a new graphic based on the graphic in the review article (see figure to the right). While the figure may not be eye-catching, I do believe it is informative. In my opinion, informative is more important than eye catching. Thoughts? Boghog (talk) 21:42, 4 February 2017 (UTC)
References
- ^ Max EE (1986). "Plagiarized Errors and Molecular Genetics". Creation Evolution Journal. 6 (3): 34–46.
- ^ Chandrasekaran C, Betrán E (2008). "Origins of new genes and pseudogenes". Nature Education. 1 (1): 181.
- I still vote for mine, but I'm open to a consensus developing against it. The reason I let it "sit" for a couple weeks without devoting the extra time to the superoxide bullet was to see if a consensus would develop against it. When I heard no complaints, I made the additional subtle change, which I think made it even better. But, I'm not going to get into an edit war about it. I have mixed emotions about eye candy in wp, and who(m) decides what is eye candy and what is significant. DennisPietras (talk) 00:57, 5 February 2017 (UTC)
- In determining consensus, strength of argument is more important than counting votes. The "shooter" in the present diagram apparently was created to represent the Green-beard effect which has little if anything to do with pseudogenes. The present diagram implies that source of reactive oxygen species is DNA itself. Apparently some photosensitizers like berberine only generate singlet oxygen when bound to DNA (PMID 22313410). However this is a very specialized reaction is only one a of large number of mechanisms that could potential generate pseudogenes. Without a reliable source that directly links this mechanism to pseudogene formation, this amounts to original research which is prohibited in Wikipedia. The figure above is much more general, is supported by reliable sources, and directly supports the text in the article. The current text says nothing about specific mechanisms of mutations nor in my view, should it since it somewhat off topic. The same can be said of the current figure. Boghog (talk) 07:39, 5 February 2017 (UTC)
- I still vote for mine, but I'm open to a consensus developing against it. The reason I let it "sit" for a couple weeks without devoting the extra time to the superoxide bullet was to see if a consensus would develop against it. When I heard no complaints, I made the additional subtle change, which I think made it even better. But, I'm not going to get into an edit war about it. I have mixed emotions about eye candy in wp, and who(m) decides what is eye candy and what is significant. DennisPietras (talk) 00:57, 5 February 2017 (UTC)
Edits February 2020[edit]
I'm doing some revisions, to polish and simplify the text and to expand the evolution information. Before deciding what to change I'm consulting with the Gerstein Lab and Larry Moran.
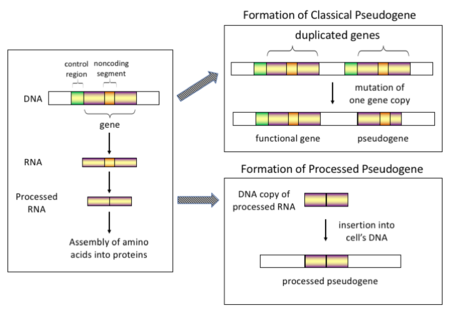
So far I've only changed the Lead section, but I plan on doing more over the next few weeks. This may have removed the need for some references; I'll go back and clean these up soon. I also moved the Pseudogene formation figure down to the Types and Origins section, and added a simpler figure in the Lead section that just shows the types of defects commonly seen in pseudogenes. Rosieredfield (talk) 22:23, 12 February 2020 (UTC)
The relation between pseudogenes and non-coding DNA[edit]
@Elemimele: Non-coding DNA is a popular term for all of the DNA that's not coding but it's not particularly useful. We could also have articles for non-centromeric DNA, non-regulatory DNA, and non-intron DNA but that would be silly, wouldn't it?. The only reason for emphasizing non-coding DNA is historical - it relates to the incorrect notion that in the 1970s the only important DNA in a genome was coding DNA.
Nevertheless, we are stuck with the term so we've dealt with it by putting brief mentions of all the different kinds of non-coding DNA in the Non-coding DNA article. However, those are just brief mentions and they link to the main articles.
This is one of the main articles. It contains a more complete discussion of pseudogenes. We do not have to refer back to all of the Wikipedia articles that mention pseudogenes and link to this main article on pseudogenes.
In addition, the recent edit deleted the description of pseudogenes as nonfunctional but that's an important part of the lead. When the new Junk DNA article is created there will be a brief summary of the reasons why pseudogenes are junk and a link to this article where there will be a more extensive discussion.
I recommend reverting the edit by Quercus solaris. Genome42 (talk) 22:50, 4 March 2023 (UTC)
- Are pseudogenes hyponymous to junk DNA, and is junk DNA hyponymous to noncoding DNA? The comment above seems to suggest that the answer is "yes and yes, but those relationships are not important, because the top hypernymous category (noncoding DNA) is so heterogeneous as to be unuseful for many purposes." OK, then what if the opening sentence of this article said, "Pseudogenes are a type of junk DNA that resembles functional genes"? Would that be a good expository sentence or not, yes or no? The fact that this article (e.g., lede, body, hatnotes), before today, neither mentioned nor linked to either noncoding DNA or junk DNA at all, in any way, is an obvious pedagogical problem. I
will go changejust changed my previous edit. If the new version is still not good, then who will explain a succinct correction that simply fixes it? A remaining expository problem in this lede is that it speaks of "functional genes." Very well, that sets up a comparison with "nonfunctional genes". What then is the definition of a "nonfunctional gene", and how is that differentiated (if indeed it is) from a pseudogene? If all genes are functional, then why are we speaking of functional genes? Wikipedia ledes want to know, whatever the answers may be. If they can't explain it, then they obviously need more expository work. Quercus solaris (talk) 05:33, 5 March 2023 (UTC)- No, Quercus solaris, I still disagree completely with Genome42's reversion, and prefer your original version. A large proportion of our readers will have a firm idea in their heads that DNA is connected with genes and genes encode proteins that do stuff, because that is what has been taught in elementary level biology for at least 30 years. They wouldn't recognise a centromere if they tripped over it. They have no idea of the wonderful world of non-coding DNA (there is a reason why that term is popular, while terms like non-centromeric etc. are not).
- Let's not allow our biology articles to descend into the mess that the mathematicians have done with their articles, where, through a failure to distinguish pedanticism from rigour, it's become almost a point of honour to be unhelpful to readers on the grounds that they ought to know what they're talking about before reading the article. We're better than that.
- Can we have the link to non-coding DNA back please? It's an important article, rather closely linked to this one in subject.
- It's also an article that's getting into a mess because of recent editing. It's now got two completely separate sections on the same subject, separated by other stuff, which is plain weird. Elemimele (talk) 10:12, 5 March 2023 (UTC)
- @Quercus solaris: Thank-you for fixing the edit. The current version is excellent.
- I had to look up the word hypernymous and found this definition: "A word whose meaning is included in the meaning of another more general word; for example, bus is a hyponym of vehicle." I'm not sure how it fits into the discussion. My view is that it's better to describe the larger category to which pseudogenes BELONG rather than the nebulous category that they don't belong to (e.g. non-coding DNA). Hence, saying that pseudogenes are not coding DNA is not helpful but describing them as a subset of junk DNA is helpful.
- The explanation of the function wars will go into the new Junk DNA article. For now, we are often stuck with making a distinction between what some people think are functional genes and what other people are calling functional genes using a non-standard definition of function.
- Some of this applies to pseudogenes since, as you probably know, there are papers out there claiming that most (all?) pseudogenes have a biologically relevant function. (Cheetham et al., 2020 - ref #25). That's going to take us down into the swamp of the function wars. Genome42 (talk) 16:47, 5 March 2023 (UTC)
- I think you're running the risk of creating a few misconceptions here. You write that non-coding DNA is a "nebulous" category, but it's not nebulous at all. It's very clear: it's all the category of all DNA that doesn't code for anything. You write that pseudogenes don't belong to this category, the category of DNA-that-doesn't-code-for-anything, but clearly the whole point of pseudogenes is that they aren't transposed, translated, and they don't make anything - i.e. they don't code for anything. They are a subset of non-coding DNA, which is why there was already a section on pseudogenes in the article on non-coding DNA.
- If you want an article on junk DNA, go ahead. It's an important concept that has existed. But you can't avoid the swamp of the function wars because whether "junk" is truly junk is a huge part of the debate on junk DNA. And the potential evolutionary usefulness of pseudogenes (and the difficulty of establishing exactly how pseudo a pseudogene is) is a huge part of that debate too. Your article will be incomplete if it doesn't cover this stuff (and I do note that you are trying to do so, which I appreciate).
- Incidentally, as someone who has worked for the last 23 years in assorted genetics facilities, it's been a good 15 years since I heard any of my colleagues refer to "junk DNA". I know this isn't relevant, as WP is neither interested in editors' personal expertise, nor vignettes from the lab. But please, as you write the article, do remember that our readers are not geneticists. To them, the revelation that DNA might not encode a gene is a bit of a surprise, and prompts the question "Well, if it's not encoding a gene, what is it doing?" We're here to answer that question. I don't mind an article on junk dna, but I don't want it to detract from the article on non-coding DNA or kidnap parts of that article, which is fulfilling a very useful role. Elemimele (talk) 17:59, 5 March 2023 (UTC)
- Hi all. Excuse the length below, but I promise it's worthwhile for this audience (of us WP editors). TL;DR—the upshot is that overall we're getting there, not too bad. In my view the current lede version (as I write this) is not great, but it is "good enough" to solve the biggest problem with the old one, and it's not inferior enough (to a better one) for me to campaign for a different one at the moment. Overall I agree with Elemimele's viewpoints above. The good news (for those viewpoints) is that the current lede version (as I write this) is "good enough" not to suck too bad, because any learner who clicks through to junk DNA (regardless of whether now, when it redirects to a section of the article on non-coding DNA, or in the future, when it may be its own article that clearly links to non-coding DNA) is going to learn pretty easily the "You Are Here" upshot on the semantic relations map (which is also the ontologic map) of "what pines and trees and plants are", so to speak (I explain this metaphor below). Regarding hyponymy and hypernymy, there is nothing complicated about the relation—it is the simply the concept that pines are a subset of trees and trees are (in turn) a subset of plants, for example. Layperson readers need exposition to tell them such basic relationships because they have no idea otherwise; it is ontological blindness: "Are all pines trees, or only 'those pines that are important enough to human interests that we care enough to talk about them'? Are all trees pines, or no? Is there any type of trees that aren't plants?" and so on. Such concepts are not complicated, and yet people (learners) need them pointed out (made clear) when they learn about technical things (e.g., molecules, devices, abstractions) because otherwise they have no clue (which is natural, not their fault). And teachers who can't clearly and simply give those cardinal/orientation clues aren't doing an adequate job of exposition. For example, there's a place in life for saying a concept like, "All splarks are squarks, but when people talk about splarks they are usually referring to the most important class of them, which is splarks that have slarks in them." Without diving into any rabbit holes of technicalities, such a sentence still makes quite clear to layperson learners the semantic/ontologic upshot. That's the goal as far as exposition and pedagogy/teaching go. (An aside about those terms: apparently nowadays some laypeople are afraid of the word pedagogy because they're too dumb to understand that it doesn't mean "pedo-you-know-whatnow". That's the level of dangerous weapons-grade ignorance that the world is up against, now more than ever.) One more thought, related to such ignorance: there is usage prescription advice out there in the world of science editing to avoid the term junk DNA because it leads laypersons to mistakenly assume that any DNA that doesn't code peptides is just "garbage". This advice wants to use the term non-coding DNA to avoid that misapprehension, but doing so may introduce another infecility, which is that the umbrella category is very wide. It would be as if someone said, "please avoid the word 'tree' because it is too confusing," but then the alternative of calling pines 'plants' strikes some initiates as too broad to be useful. A saving grace (for those who worry about finding an optimal solution) is that, at the end of the day, it's not super critical which term Wikipedia uses in such a case, as long as it's not frankly racist, denotatively distorted/wrong (as opposed to merely connotatively inoptimal), or similarly horrible. And when we keep in mind that Wikipedia will never be the last word (i.e., learners who need more can turn to non-crowdsourced science publications in addition), we can be OK with Wikipedia not necessarily doing it perfectly, as long as it is pretty good and good enough. And thank you for giving a shit either way—because even that alone is more than most people bother to do. Cheers, Quercus solaris (talk) 19:49, 5 March 2023 (UTC)
- I enjoyed your explanation about pines, "Regarding hyponymy and hypernymy, there is nothing complicated about the relation—it is the simply the concept that pines are a subset of trees and trees are (in turn) a subset of plants, for example." But you forgot to mention that pines are also "not animals" and "not bacteria" and that's the problem. In an article on pines do we really need to explain in the lead that pines are not animals and not bacteria? Of course not, that would just be confusing.
- Genomes are divided into many different categories. For example, they could be split into functional regions and non-functional regions (junk) and pseudogenes would clearly fall into the later category. That's a much more logical split than coding and non-coding or centromeres and non-centromeres but it's also a sort of "this and not this" split. (Nobody said that biology is easy.)
- You are correct to assume that the root of the problem is the widespread, and incorrect, belief that the term "junk DNA" originally referred to all non-coding DNA. That false narrative is so pervasive among the general public that they are convinced that the discovery of functional DNA in the noncoding part of the genome has completely refuted the concept of junk DNA.
- We will have to work hard to correct those sorts of misconceptions and it's going to be difficult since they are also widespread among Wikipedia editors. I'm glad to see that you are willing to help fix the problem. Genome42 (talk) 20:18, 5 March 2023 (UTC)
- Hi all. Excuse the length below, but I promise it's worthwhile for this audience (of us WP editors). TL;DR—the upshot is that overall we're getting there, not too bad. In my view the current lede version (as I write this) is not great, but it is "good enough" to solve the biggest problem with the old one, and it's not inferior enough (to a better one) for me to campaign for a different one at the moment. Overall I agree with Elemimele's viewpoints above. The good news (for those viewpoints) is that the current lede version (as I write this) is "good enough" not to suck too bad, because any learner who clicks through to junk DNA (regardless of whether now, when it redirects to a section of the article on non-coding DNA, or in the future, when it may be its own article that clearly links to non-coding DNA) is going to learn pretty easily the "You Are Here" upshot on the semantic relations map (which is also the ontologic map) of "what pines and trees and plants are", so to speak (I explain this metaphor below). Regarding hyponymy and hypernymy, there is nothing complicated about the relation—it is the simply the concept that pines are a subset of trees and trees are (in turn) a subset of plants, for example. Layperson readers need exposition to tell them such basic relationships because they have no idea otherwise; it is ontological blindness: "Are all pines trees, or only 'those pines that are important enough to human interests that we care enough to talk about them'? Are all trees pines, or no? Is there any type of trees that aren't plants?" and so on. Such concepts are not complicated, and yet people (learners) need them pointed out (made clear) when they learn about technical things (e.g., molecules, devices, abstractions) because otherwise they have no clue (which is natural, not their fault). And teachers who can't clearly and simply give those cardinal/orientation clues aren't doing an adequate job of exposition. For example, there's a place in life for saying a concept like, "All splarks are squarks, but when people talk about splarks they are usually referring to the most important class of them, which is splarks that have slarks in them." Without diving into any rabbit holes of technicalities, such a sentence still makes quite clear to layperson learners the semantic/ontologic upshot. That's the goal as far as exposition and pedagogy/teaching go. (An aside about those terms: apparently nowadays some laypeople are afraid of the word pedagogy because they're too dumb to understand that it doesn't mean "pedo-you-know-whatnow". That's the level of dangerous weapons-grade ignorance that the world is up against, now more than ever.) One more thought, related to such ignorance: there is usage prescription advice out there in the world of science editing to avoid the term junk DNA because it leads laypersons to mistakenly assume that any DNA that doesn't code peptides is just "garbage". This advice wants to use the term non-coding DNA to avoid that misapprehension, but doing so may introduce another infecility, which is that the umbrella category is very wide. It would be as if someone said, "please avoid the word 'tree' because it is too confusing," but then the alternative of calling pines 'plants' strikes some initiates as too broad to be useful. A saving grace (for those who worry about finding an optimal solution) is that, at the end of the day, it's not super critical which term Wikipedia uses in such a case, as long as it's not frankly racist, denotatively distorted/wrong (as opposed to merely connotatively inoptimal), or similarly horrible. And when we keep in mind that Wikipedia will never be the last word (i.e., learners who need more can turn to non-crowdsourced science publications in addition), we can be OK with Wikipedia not necessarily doing it perfectly, as long as it is pretty good and good enough. And thank you for giving a shit either way—because even that alone is more than most people bother to do. Cheers, Quercus solaris (talk) 19:49, 5 March 2023 (UTC)
- @Elemimele: There's no user page for Elemimele but there's a hint that it might be a Wikipedia administrator who's also a scientist. Is this correct? If so, you could be a tremendous help in trying to fix science articles because administrators have a lot of power.
- I understand your perspective on non-coding DNA because it's very common. But part of the problem is that it has become a loaded word that carries much more meaning than just DNA that's not coding for protein. Let's try not to inject that POV into other articles unless we can't avoid it.
- I'm not surprised to hear that you've been associating with scientists who dismiss junk DNA. That's part of the problem. The true experts in this field are the scientists who specialize in molecular evolution and they are almost unanimous in declaring that most of the human genome is junk. It's a shame that this perspective has been kept off Wikipedia for many years, ever since the original Junk DNA article was deleted in 2007. It's even more of a shame that there are so many scientists who don't even know that there's an active scientific controversy that has not been settled.
- I agree with you that we are here to explain to the general reader what sorts of things are in your genome. Let's try and do as good a job of conveying accurate scientific information as we possibly can. As part of that goal, we need to explain why so many people are confused about the distinction between non-coding DNA and junk DNA. Can you help?
- We've also been working on the Gene article in order to explain the correct scientific definition of "gene" and let people know that genes occupy more that 40% of the human genome. Any help on that article will also be greatly appreciated.
- Please add your recommendation to the discussion about splitting the Non-coding DNA article and creating a separate Junk DNA article. If you are an administrator, as I suspect, your support would be greatly appreciated.
- It won't be hard to find my email address. If you send me an email I'll give you some information that you might find very helpful in sorting out this mess. Genome42 (talk) 19:57, 5 March 2023 (UTC)
- Hi Genome42, just to clear things up, I'm not an admin, and in any case admins have no special powers over content here, so your word is as good as anyone's on how to arrange the material. My background is in plants and microbes, not in humans, and I think attitudes to junk DNA are perhaps a bit different in different organisms. For example, humans can walk away from a threat, while plants and microbes tend to indulge in chemical warfare instead, which means they have, maybe, a stronger evolutionary pressure to come up with novel molecules - this means that having a resource of non-functional pseudogenes just waiting to find a function might be a benefit to a plant or microbe. In effect, it's true that a vast proportion of the genome is entirely unnecessary ("junk", doing nothing) in the sense that you could eliminate it and the plant would grow fine, it's still serving an evolutionary function in that the plant would maybe do less well over the next million years than its neighbour because it lacks the mechanism to do anything new. When genes duplicate, their products often diverge in function.
- My feeling is that an article on junk DNA does make sense, and the more I read your arguments, the more I think you're absolutely right to write one. I think the junk DNA article should give an overview of the big debate on how much of the genome (in various organisms) is doing something "useful", and should indeed describe the arguments biologists have had, to decide when DNA is genuinely "junk", and its effects on the evolutionary path of an organism. It should also cover the history of the term, because that reflects the history of biological thinking, and as an encyclopaedia, we have to tell the history as well as the current beliefs (we are read by the general public, including both biologists and historians). I'm grateful you've put so much effort into tracing just when the term came into use. "Junk" is itself a loaded term, but the loading is part of the development of evolutionary biology.
- I think it's also valuable to have the non-coding DNA article, and it's quite reasonable for the two to exist in parallel, and for each to refer to the other. The non-coding DNA article can list pseudogenes, but the junk DNA article is a very valuable addition as it discusses not so much a class of DNA in the sense of "centromeres", "genes" etc., but a past and current biological debate about the usefulness of large chunks of the genome. Why they're there, and what they're doing (if anything). I definitely don't think the subject should be suppressed.
- There are loads of good refs, obviously.
- On the >40% thing, I am not a human geneticist, but you're going to struggle to get that value fully accepted without clarification and explanation, because here people will bump into reputable sources giving quite different values (e.g. here: [1] The problem with Wikipedia is that it doesn't particularly care what is right or what is the latest thinking, it cares only about summarising what "reliable sources" have said. In situations where apparently reliable sources have said what looks like one thing, and you want to argue for another, the best thing to do is probably include both, and a balanced discussion of why the values differ.
- It's possible we'll argue over content, but I am convinced that junk DNA as a topic is an encyclopaedic one, and valuable. Elemimele (talk) 11:11, 6 March 2023 (UTC)
- @ Elemimele: There's a lot of confusion about your identity on your talk page. Thanks for giving me a bit of background.
- Since you support the creation of a separate talk page, I'd appreciate it if you would voice that support in the Talk section of the Non-coding DNA article.
- I understand the teleological argument for junk DNA but I'm not sure it can be covered effectively in the new article. It should be covered under species selection somewhere. The current model for the existence of junk DNA is related to nearly neutral theory and population size and not some intrinsic (and future) benefit of having junk DNA.
- You bring up a very interesting point about how to define a gene. It's covered in Gene but many readers aren't going to bother reading that article since they think they know what a gene is. You are correct to point out that there are many references in the scientific literature that say genes only occupy 1% of the human genome. All of these make the same mistake of equating coding regions with genes.
- In order for the 1% claim to be true they must be assuming two things: (1) protein-coding genes are the only type of gene, and (2) gene is defined as only the coding region of a protein-coding gene and not the introns or the 5' and 3' UTRs. That conflicts with the definition we have described in the Gene article.
- In the real world we wouldn't have a problem sorting this out since all reasonable scientists would see the problem and realize that their colleagues are wrong to say that genes only occupy 1% of the genome. But on Wikipedia this is a problem. As you point out, Wikipedia editors don't care about what is correct or reasonable. Their goal is to document and publicize all the misinformation that's out there as long as it comes from a "reliable source." As you say, the only way to resolve issues like this are to document conflicting "reliable sources" that say 40% and 1% and explain the difference without being accused of editorializing. That's extremely frustrating.
- The Wikipedia culture is set up to resist any correcting of misinformation. Some of us are interested in changing the culture so that the focus is on facts and not process. Please help. Genome42 (talk) 18:35, 6 March 2023 (UTC)
- Wikipedia requested biology diagrams
- B-Class Molecular Biology articles
- Unknown-importance Molecular Biology articles
- B-Class Genetics articles
- High-importance Genetics articles
- WikiProject Genetics articles
- B-Class MCB articles
- Mid-importance MCB articles
- WikiProject Molecular and Cellular Biology articles
- All WikiProject Molecular Biology articles



