C2orf72
| C2orf72 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | C2orf72, chromosome 2 open reading frame 72, Chromosome 2, Open Reading Frame 72 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1920042 HomoloGene: 54780 GeneCards: C2orf72 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
C2orf72 (Chromosome 2, Open Reading Frame 72) is a gene in humans (Homo sapiens) that encodes a protein currently named after its gene, C2orf72.[5] It is also designated LOC257407[5] and can be found under GenBank accession code NM_001144994.2.[6] The protein can be found under UniProt accession code A6NCS6.[7]
This gene is primarily expressed in the liver, brain, placental, and small intestine tissues.[8] C2orf72 is an intracellular protein that has been predicted to reside within the nucleus, cytosol, and plasma membrane of cells.[5] The function of C2orf72 is unknown, but it is predicted to be involved in very-low-density lipoprotein particle assembly and also involved in the regulation of cholesterol esterification.[9] This prediction also matches with the fact that both estradiol[10] and testosterone[11] have been reported to upregulate expression of C2orf72.[12]
Gene[edit]


Locus[edit]
C2orf72 is a protein-coding gene found on the forward (+) strand of chromosome 2 at the locus 2q37.1, on the long arm of the chromosome.[5]
mRNA[edit]
C2orf72's mRNA transcript is reported to be about 3,629 base pairs long.[6] It appears to have two polyadenylation sites near the 5′ end of the mRNA transcript, each preceded by their respective regulatory sequences, such as ATTAAA or AATAAA.[6]
There are three predicted exons reported for human C2orf72.[6]
Expression pattern[edit]
C2orf72 is preferentially expressed in brain, liver, placenta, colon, small intestine, gallbladder, stomach, and prostate, and to a lesser extent in adrenal gland, appendix, pancreas, lung, kidney, testis, and urinary bladder.[8]
Predicted Biological Functions[edit]

It is predicted via Archs4[13] (July 16, 2022) that the function of this gene may be related to very-low-density lipoprotein particle assembly[14] and also involved in the regulation of cholesterol esterification.[9]
Regulation[edit]
Gene-level regulation[edit]
Gene perturbation data[edit]
In a study of embryonic liver samples lacking hepatocyte nuclear factor 4 alpha (HNF4α), the expression of C2orf72 was downregulated.[15]
Both estradiol[10] and testosterone[11] upregulate expression of C2orf72.[12]
Expression pattern[edit]
C2orf72 mRNA and protein products are found preferentially in the liver, kidney, and placenta.[16] The protein is localized to the cell membrane and cytoplasm in liver, brain, and placental tissues.[16]
Transcript-level regulation[edit]
miR-1271-5p is a microRNA that could bind to the 3′ untranslated region of the C2orf72 mRNA transcript at 5′-...GUGCCAA...-3′.[6][17][18]
Protein-level regulation[edit]
Predicted phosphorylation sites[edit]
There are at least two predicted phosphorylation sites for the human C2orf72 protein, one at threonine-286 and the other at serine-294.[7]

Protein[edit]




Human protein[edit]
The predicted molecular weight of C2orf72 is 30.5 kDa,[19] and it has a predicted isoelectric point (pI) of pH 8.7.[20]
There are eight cysteine residues, for a potential of four disulfide bonds.[21] Most of the cysteine residues are positioned next to a polar amino acid (uncharged or positively or negatively charged).[21]
At physiological pH, there are 33 positively charged amino acid residues, including histidine, most of which are arginines.[21] Likewise, there are 33 negatively charged amino acid residues, most of which are glutamates.[21]
There are 14 hydroxyl-containing residues (tyrosine, threonine or serine) that could serve as typical phosphorylation sites; most of these are serines.[21]
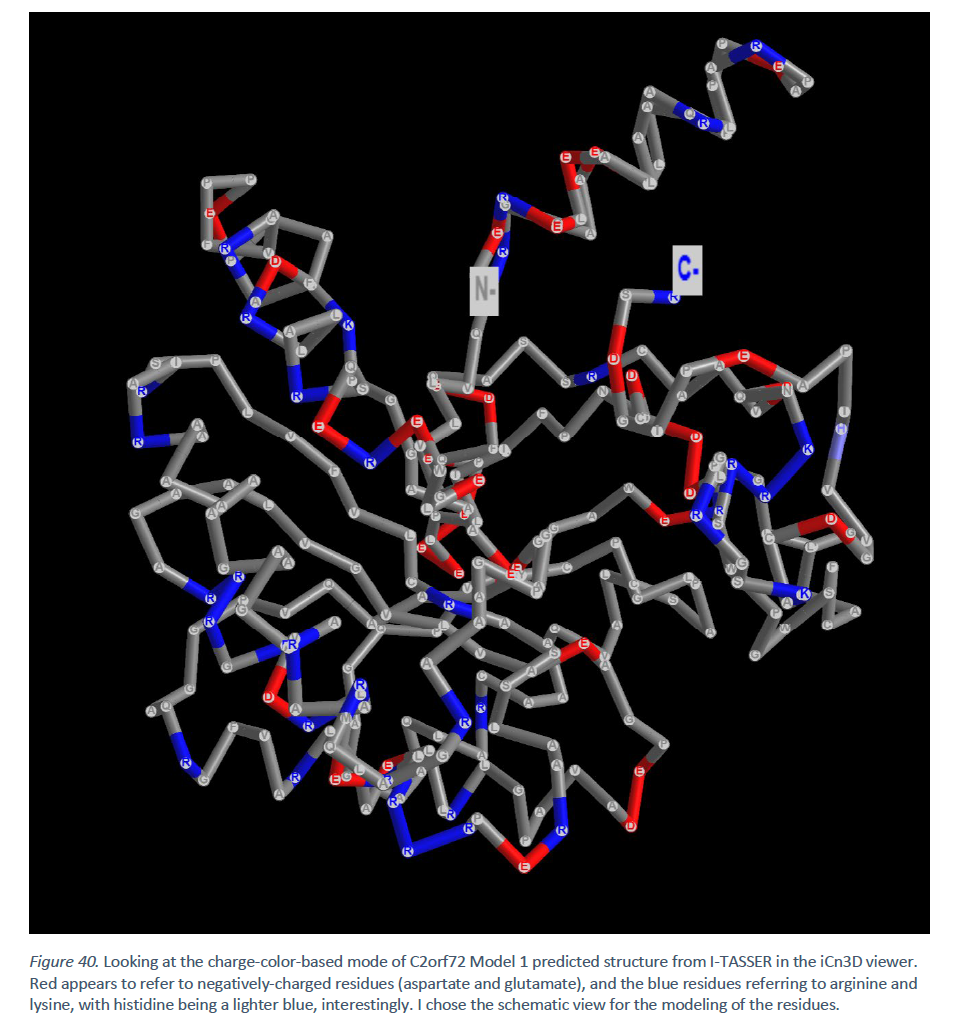
Interacting proteins[edit]
These proteins have been reported to interact with human C2orf72: RASN (GTPase NRas),[23] RASK (GTPase KRas),[23] and CD81.[24][25]
Homology[edit]
There are at least 203 organisms with an ortholog of C2orf72.[26] The most evolutionarily distant reported ortholog of C2orf72 is in the Australian ghost shark (Callorhincus milii);[27][28][29], and it is broadly conserved from Actinopterygii (bony fish) to Mammalia.
| Genus and species | Common name | Order | Date of divergence from human
(million years ago) |
GenBank accession
code |
Sequence
length |
Sequence identity (%) | Sequence
similarity (%) |
|---|---|---|---|---|---|---|---|
| Pan troglodytes | Chimpanzee | Primates | 6.7 | XP_516141.5 | 295 | 98.6 | 98.6 |
| Pongo abelii | Sumatran orangutan | Primates | 15.76 | XP_024099683.1 | 295 | 95.3 | 96.9 |
| Castor canadensis | American beaver | Rodentia | 90 | XP_020011841.1 | 282 | 77.6 | 82.4 |
| Oryx dammah | Scimitar-horned oryx | Artiodactyla | 96 | XP_040084064.1 | 285 | 74.6 | 79.3 |
| Sus scrofa | Wild boar | Artiodactyla | 96 | XP_005657646.1 | 282 | 75.3 | 80.7 |
| Tursiops truncatus | Common bottlenose dolphin | Cetacea | 96 | XP_033715450.1 | 285 | 76.9 | 80.7 |
| Felis catus | Domestic cat | Carnivora | 96 | XP_023115562.1 | 286 | 80.1 | 83.1 |
| Eptesicus fuscus | Big brown bat | Chiroptera | 96 | XP_027993078.1 | 151 | 36.1 | 38.9 |
| Corapipo altera | White-ruffed manakin | Passeriformes | 312 | XP_027503457.1 | 181 | 26.7 | 34.0 |
| Pipra filicauda | Wire-tailed manakin | Passeriformes | 312 | XP_027606890.1 | 243 | 34.7 | 45.2 |
| Taeniopygia guttata | Zebra finch | Passeriformes | 312 | XP_030136117.3 | 255 | 35.1 | 45.4 |
| Corvus cornix cornix | Hooded crow | Passeriformes | 312 | XP_039412719.1 | 245 | 36.0 | 45.3 |
| Hirundo rustica | Barn swallow | Passeriformes | 312 | XP_039930397.1 | 243 | 37.0 | 46.7 |
| Aythya fuligula | Tufted duck | Anseriformes | 312 | XP_032049188 | 251 | 36.3 | 46.7 |
| Anas platyrhynchos | Mallard | Anseriformes | 312 | XP_038039556.1 | 251 | 36.3 | 46.7 |
| Protobothrops mucrosquamatus | Brown-spotted pit viper | Squamata | 312 | XP_029139335.1 | 278 | 22.9 | 34.5 |
| Python bivittatus | Burmese python | Squamata | 312 | XP_025023716.1 | 279 | 23.3 | 35.9 |
| Pseudonaja textilis | Eastern brown snake | Squamata | 312 | XP_026577460.1 | 272 | 31.6 | 41.0 |
| Pantherophis guttatus | Corn snake | Squamata | 312 | XP_034263860.1 | 252 | 33.0 | 42.5 |
| Pogona vitticeps | Central bearded dragon | Squamata | 312 | XP_020657305.1 | 295 | 24.1 | 34.0 |
| Zootoca vivipara | Common lizard | Squamata | 312 | XP_034989711.1 | 285 | 37.9 | 48.6 |
| Lacerta agilis | Sand lizard | Squamata | 312 | XP_033004091.1 | 289 | 38.0 | 49.5 |
| Podarcis muralis | Common wall lizard | Squamata | 312 | XP_028587763.1 | 272 | 38.7 | 50.8 |
| Gopherus evgoodei | Goode's thornscrub tortoise | Testudines | 312 | XP_030431493.1 | 481 | 24.2 | 31.1 |
| Terrapene carolina
triunguis |
Three-toed box turtle | Testudines | 312 | XP_029766982.1 | 262 | 35.1 | 43.2 |
| Chrysemys picta bellii | Painted turtle | Testudines | 312 | XP_023966073.1 | 306 | 36.6 | 47.4 |
| Dermochelys coriacea | Leatherback sea turtle | Testudines | 312 | XP_038272534.1 | 271 | 38.1 | 48.1 |
| Mauremys reevesii | Reeves' turtle | Testudines | 312 | XP_039344659.1 | 277 | 39.5 | 51.4 |
| Nanorana parkeri | High Himalaya frog | Anura | 351.8 | XP_018432004.1 | 304 | 27.3 | 40.1 |
| Xenopus tropicalis | Tropical clawed frog | Anura | 351.8 | XP_002937397.3 | 289 | 30.7 | 42.4 |
| Rhinatrema bivittatum | Two-lined caecilian | Gymnophiona | 351.8 | XP_029473197.1 | 358 | 30.3 | 36.1 |
| Geotrypetes seraphini | Gaboon caecilian | Gymnophiona | 351.8 | XP_033814148.1 | 233 | 33.9 | 44.2 |
| Parambassis ranga | Indian glass fish | Perciformes | 435 | XP_028260036.1 | 334 | 19.7 | 34.5 |
| Acanthochromis polyacanthus | Spiny chromis | Perciformes | 435 | XP_022050415.1 | 317 | 21.8 | 35.6 |
| Acanthopagrus latus | Yellowfin seabream | Perciformes | 435 | XP_036971960.1 | 309 | 22.0 | 35.5 |
| Cyprinodon tularosa | White Sands pupfish | Cyprinodontiformes | 435 | XP_038147473.1 | 296 | 20.1 | 33.1 |
| Esox lucius | Northern pike | Esociformes | 435 | XP_012990404.1 | 332 | 20.6 | 33.1 |
| Thunnus maccoyii | Southern bluefin tuna | Scombriformes | 435 | XP_042273029.1 | 329 | 20.2 | 34.0 |
| Syngnathus acus | Greater pipefish | Syngnathiformes | 435 | XP_037106050.1 | 274 | 19.5 | 34.9 |
| Callorhinchus milii | Australian ghost shark | Chimaeriformes | 473 | XP_007887618.1 | 413 | 17.6 | 26.5 |
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000204128 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000026227 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d "C2orf72 GeneCards". www.genecards.org. Retrieved 2021-08-02.
- ^ a b c d e "Homo sapiens chromosome 2 open reading frame 72 (C2orf72), mRNA". 2020-12-12 – via NCBI Nucleotide.
- ^ a b "iPTMnet Report A6NCS6 C2orf72". research.bioinformatics.udel.edu. Retrieved 2021-08-02.
- ^ a b "C2orf72 chromosome 2 open reading frame 72 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-08-02.
- ^ a b "ARCHS4". maayanlab.cloud. Retrieved 2021-08-02.
- ^ a b "Gene Set - estradiol_homo sapiens_gpl570_gds3283". maayanlab.cloud. Retrieved 2021-08-02.
- ^ a b "Gene Set - testosterone_mus musculus_gpl1261_gse17553". maayanlab.cloud. Retrieved 2021-08-02.
- ^ a b "Gene - C2ORF72". maayanlab.cloud. Retrieved 2021-08-02.
- ^ "ARCHS4". maayanlab.cloud. Retrieved 2022-07-16.
- ^ "QuickGO". www.ebi.ac.uk. Retrieved 2022-07-16.
- ^ "Gene Set - hnf4a_16714383_e18dot5_liver_lof_mouse_gpl1261_gds1916". maayanlab.cloud. Retrieved 2021-08-02.
- ^ a b "Tissue expression of C2orf72 - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2021-08-02.
- ^ "miRDB - MicroRNA Target Prediction Database". mirdb.org. Retrieved 2021-08-02.
- ^ "TargetScanHuman 7.2". www.targetscan.org. Retrieved 2021-08-02.
- ^ "C2orf72 protein expression summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2021-08-02.
- ^ "Compute pI/MW - SIB Swiss Institute of Bioinformatics | Expasy". www.expasy.org. Retrieved 2021-08-02.
- ^ a b c d e "uncharacterized protein C2orf72 [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-08-02.
- ^ "I-TASSER server for protein structure and function prediction". zhanggroup.org. Retrieved 2022-07-16.
- ^ a b Kovalski JR, Bhaduri A, Zehnder AM, Neela PH, Che Y, Wozniak GG, Khavari PA (February 2019). "The Functional Proximal Proteome of Oncogenic Ras Includes mTORC2". Molecular Cell. 73 (4): 830–844.e12. doi:10.1016/j.molcel.2018.12.001. PMC 6386588. PMID 30639242.
- ^ Bruening J, Lasswitz L, Banse P, Kahl S, Marinach C, Vondran FW, et al. (July 2018). "Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB". PLOS Pathogens. 14 (7): e1007111. doi:10.1371/journal.ppat.1007111. PMC 6053247. PMID 30024968.
- ^ "HitPredict - High confidence protein-protein interactions". www.hitpredict.org. Retrieved 2021-08-02.
- ^ "C2orf72 orthologs". NCBI. Retrieved 2021-08-04.
- ^ "LOC103176070 uncharacterized protein C2orf72 homolog [Callorhinchus milii (elephant shark)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-08-04.
- ^ "uncharacterized protein C2orf72 homolog [Callorhinchus milii] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-08-08.
- ^ "PREDICTED: uncharacterized protein C2orf72 homolog Callorhinchus mili - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2021-08-08.




